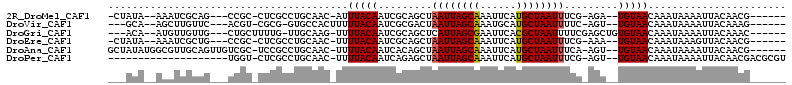
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,233,910 – 12,234,006 |
| Length | 96 |
| Max. P | 0.992431 |

| Location | 12,233,910 – 12,234,006 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -14.75 |
| Energy contribution | -14.67 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
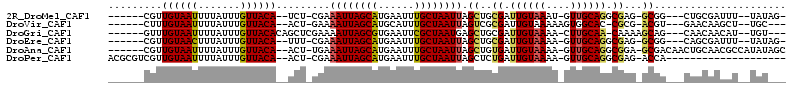
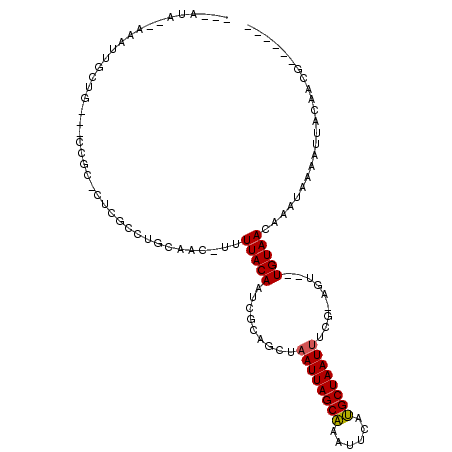
>2R_DroMel_CAF1 12233910 96 + 20766785 ------CGUUGUAAUUUUAUUUGUUACA--UCU-CGAAAUUAGCAUGAAUUUGCUAAUUAGCUGCGAUUGUAAAU-GUUGCAGGCGAG-GCGG---CUGCGAUUU--UAUAG- ------(((((((((.......))))))--.((-((.((((((((......))))))))..(((((((.......-))))))).))))-))).---.........--.....- ( -25.80) >DroVir_CAF1 119241 94 + 1 ------CUUUGUAAUUUUAUUUGUUACA--ACU-GAAAAUUAGCAUGCAUUUGCUAAUUAGUCGCGAUUGUAAAAAGUGGCAC-CGCG-ACGU---GAACAAGCU--UGC--- ------.............((((((...--((.-...((((((((......)))))))).((((((..(((........))).-))))-))))---.))))))..--...--- ( -23.50) >DroGri_CAF1 120706 97 + 1 ------GUUUGUAAUUUUAUUUGUUACACAGCUCGAAAAUUAGCGUGAAUUCGCUAAUGAGCUGCGAUUGUAAAA-CUUGCAA-CAAAAGCAG---CAACAACAU--UGU--- ------(((((((((.......)))))).)))......(((((((......)))))))..(((((..((((....-......)-)))..))))---)........--...--- ( -23.10) >DroEre_CAF1 102788 96 + 1 ------CGUUGUAACUUUAUUUGUUACA--UUU-CGAAAUUAGCAUGAAUUUGCUAAUUAGCUGCGAUUGUAAAA-GUUGCAGGCGAG-GCGG---CAGCGAUUU--UAUAG- ------((((((..((((..........--...-...((((((((......))))))))..((((((((.....)-)))))))..)))-)..)---)))))....--.....- ( -27.90) >DroAna_CAF1 87860 102 + 1 ------CGUUGUAAUUUUAUUUGUUACA--ACU-UGAAAUUAGCAUGAAUUUGCUAAUUAGCUGUGAUUGUAAAA-GUUGCAGGCGGA-GCGACAACUGCAACGCCAUAUAGC ------.((((((((.......))))))--)).-...((((((((......)))))))).((((((..((((...-..))))((((..-(((.....)))..)))).)))))) ( -32.30) >DroPer_CAF1 98952 88 + 1 ACGCGUCGUUGUAAUUUUAUUUGUUACA--ACU-CGAAAUUAGCAUGAAUUUGCUAAUUAGCUCUGAUUGUAAAA-GUUGCAGGCGAG-ACCA-------------------- ..(.(((((((((((.......))))))--))(-(..((((((((......)))))))).((.(((.((....))-....))))))))-))).-------------------- ( -22.90) >consensus ______CGUUGUAAUUUUAUUUGUUACA__ACU_CGAAAUUAGCAUGAAUUUGCUAAUUAGCUGCGAUUGUAAAA_GUUGCAGGCGAG_GCGG___CAGCAACUU__UAU___ .........((((((.......)))))).........((((((((......)))))))).((..((.((((((....)))))).))...))...................... (-14.75 = -14.67 + -0.08)
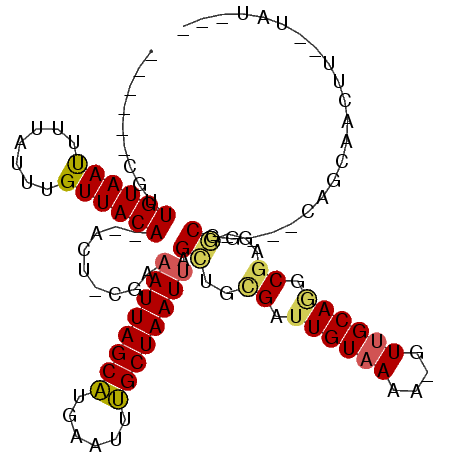
| Location | 12,233,910 – 12,234,006 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -10.45 |
| Energy contribution | -10.34 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12233910 96 - 20766785 -CUAUA--AAAUCGCAG---CCGC-CUCGCCUGCAAC-AUUUACAAUCGCAGCUAAUUAGCAAAUUCAUGCUAAUUUCG-AGA--UGUAACAAAUAAAAUUACAACG------ -.....--.....((((---.((.-..)).))))...-..(((((.(((.....((((((((......)))))))).))-)..--))))).................------ ( -17.60) >DroVir_CAF1 119241 94 - 1 ---GCA--AGCUUGUUC---ACGU-CGCG-GUGCCACUUUUUACAAUCGCGACUAAUUAGCAAAUGCAUGCUAAUUUUC-AGU--UGUAACAAAUAAAAUUACAAAG------ ---...--...(((((.---((((-((((-((.............)))))))).((((((((......))))))))...-...--.)))))))..............------ ( -21.62) >DroGri_CAF1 120706 97 - 1 ---ACA--AUGUUGUUG---CUGCUUUUG-UUGCAAG-UUUUACAAUCGCAGCUCAUUAGCGAAUUCACGCUAAUUUUCGAGCUGUGUAACAAAUAAAAUUACAAAC------ ---...--.((((((((---(.((....)-).)))).-.........(((((((((((((((......)))))))....)))))))))))))...............------ ( -26.70) >DroEre_CAF1 102788 96 - 1 -CUAUA--AAAUCGCUG---CCGC-CUCGCCUGCAAC-UUUUACAAUCGCAGCUAAUUAGCAAAUUCAUGCUAAUUUCG-AAA--UGUAACAAAUAAAGUUACAACG------ -.....--...((((((---(...-............-..........))))).((((((((......))))))))..)-)..--((((((.......))))))...------ ( -17.71) >DroAna_CAF1 87860 102 - 1 GCUAUAUGGCGUUGCAGUUGUCGC-UCCGCCUGCAAC-UUUUACAAUCACAGCUAAUUAGCAAAUUCAUGCUAAUUUCA-AGU--UGUAACAAAUAAAAUUACAACG------ (((....)))(((((((.((....-..)).)))))))-..........((((((((((((((......))))))))...-)))--)))...................------ ( -24.80) >DroPer_CAF1 98952 88 - 1 --------------------UGGU-CUCGCCUGCAAC-UUUUACAAUCAGAGCUAAUUAGCAAAUUCAUGCUAAUUUCG-AGU--UGUAACAAAUAAAAUUACAACGACGCGU --------------------..((-((((((((....-.........))).)).((((((((......))))))))..)-)((--(((((.........)))))))))).... ( -17.42) >consensus ___AUA__AAAUUGCUG___CCGC_CUCGCCUGCAAC_UUUUACAAUCGCAGCUAAUUAGCAAAUUCAUGCUAAUUUCG_AGU__UGUAACAAAUAAAAUUACAACG______ ........................................(((((.........((((((((......)))))))).........)))))....................... (-10.45 = -10.34 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:48 2006