
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,218,068 – 12,218,211 |
| Length | 143 |
| Max. P | 0.972696 |

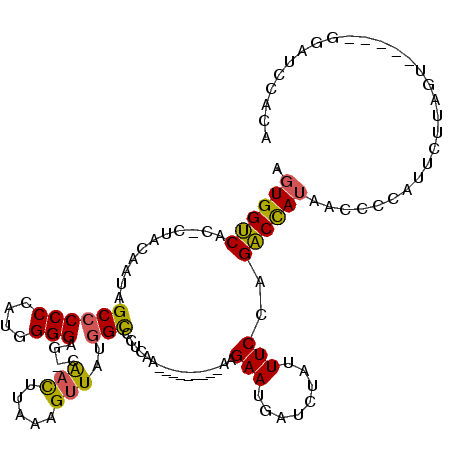
| Location | 12,218,068 – 12,218,171 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -14.15 |
| Energy contribution | -13.82 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12218068 103 + 20766785 AGUGGUCAC-CUUCAAUAGCCCCCCAUGGGGAG-UAACUUAAAGUUAUGGCCCUCGA---------AAGAAUGAUCUAUUUCCAGACCAUAGCCCCAUUCUUAGU-----UGAUCCACA .(((((((.-((......((((((....))).(-((((.....))))))))......---------(((((((..((((.........))))...))))))))).-----))).)))). ( -26.10) >DroSec_CAF1 79084 103 + 1 AGUGGUCAC-CUUCAAUAGCCCCCCAUGGGGAG-CAACUUAAAGUUAUGGCCCUCGA---------AAGAAUGAUCUAUUUCCAGACCAUAACCCCAUUCUUAGU-----UGAUCCACA .(((((((.-((......((((((...)))).)-)........(((((((...((..---------..))....(((......)))))))))).........)).-----))).)))). ( -27.90) >DroSim_CAF1 75515 103 + 1 AGUGGUCAC-CUUCAAUAGCCCCCCAUGGGGAG-CAACUUAAAGUUAUGGCCCUCGA---------AAGAAUGAUCUAUUUCCAGACCAUAACCCCAUUCUUAGU-----UGAUCCACA .(((((((.-((......((((((...)))).)-)........(((((((...((..---------..))....(((......)))))))))).........)).-----))).)))). ( -27.90) >DroEre_CAF1 87132 106 + 1 AGUGGUCACACUACAAUAGCCCCCCAUCGGGAG-CAACUUAAAGUUAUGGCCCUUAA---------AAGAAUGAUCUAUUUCCAGACCAUAACCCCAUUCUCGAU---CUGGAUCCACA .((((((.((........((.(((....))).)-)........((((((((....((---------.(((....))).))....).)))))))............---.)))).)))). ( -21.90) >DroYak_CAF1 82092 106 + 1 AGUGGUCACGCUACAAUAGCCCCCCAUCGGGAG-CAACCUAAAGUUAUGGCCCUCAG---------AAGAAUGAUCUAUUUCCAGACCAUAACCCCAUUCUUGAU---CUGGAACCACA .(((((..((...(((..((.(((....))).)-)........(((((((...(((.---------.....)))(((......)))))))))).......)))..---.))..))))). ( -24.50) >DroAna_CAF1 73848 113 + 1 AGUGGGC---UAACAAUAGCCCCCCAUUGGGAAUCGUUUUCGAGUUAUAGUCCUCACAUGGGGGCCAAGAAUGAUCUAUUUCUGGUCUAGA---CCAGUCUCAUCCAAAAGAAUUCUCU ..(((((---((....))))((((((((((((.(((....))).......))).)).))))))))))(((((.........(((((....)---))))(((........)))))))).. ( -30.70) >consensus AGUGGUCAC_CUACAAUAGCCCCCCAUGGGGAG_CAACUUAAAGUUAUGGCCCUCAA_________AAGAAUGAUCUAUUUCCAGACCAUAACCCCAUUCUUAGU_____GGAUCCACA .((((((...........((((((....)))....(((.....)))..))).................(((........)))..))))))............................. (-14.15 = -13.82 + -0.33)


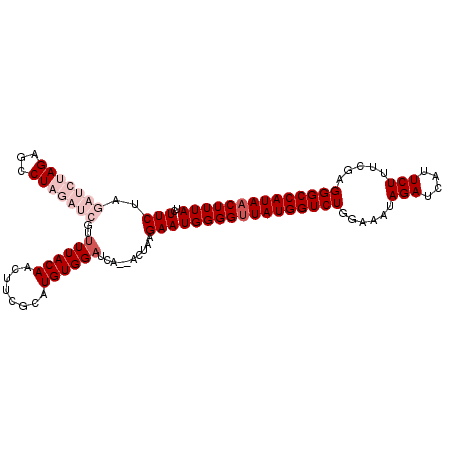
| Location | 12,218,068 – 12,218,171 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -23.09 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12218068 103 - 20766785 UGUGGAUCA-----ACUAAGAAUGGGGCUAUGGUCUGGAAAUAGAUCAUUCUU---------UCGAGGGCCAUAACUUUAAGUUA-CUCCCCAUGGGGGGCUAUUGAAG-GUGACCACU .((((.(((-----...............((((((((....)))))))).(((---------((((.((((.((((.....))))-..(((....))))))).))))))-)))))))). ( -34.00) >DroSec_CAF1 79084 103 - 1 UGUGGAUCA-----ACUAAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUU---------UCGAGGGCCAUAACUUUAAGUUG-CUCCCCAUGGGGGGCUAUUGAAG-GUGACCACU .((((.(((-----.((.....((((((((((((((.(((..(((....))))---------))..))))))))))))))(((.(-(((((....)))))).)))..))-.))))))). ( -40.50) >DroSim_CAF1 75515 103 - 1 UGUGGAUCA-----ACUAAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUU---------UCGAGGGCCAUAACUUUAAGUUG-CUCCCCAUGGGGGGCUAUUGAAG-GUGACCACU .((((.(((-----.((.....((((((((((((((.(((..(((....))))---------))..))))))))))))))(((.(-(((((....)))))).)))..))-.))))))). ( -40.50) >DroEre_CAF1 87132 106 - 1 UGUGGAUCCAG---AUCGAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUU---------UUAAGGGCCAUAACUUUAAGUUG-CUCCCGAUGGGGGGCUAUUGUAGUGUGACCACU .((((.((((.---..(((...((((((((((((((..(((.(((....))))---------))..))))))))))))))....(-(((((....))))))..)))...)).)))))). ( -38.60) >DroYak_CAF1 82092 106 - 1 UGUGGUUCCAG---AUCAAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUU---------CUGAGGGCCAUAACUUUAGGUUG-CUCCCGAUGGGGGGCUAUUGUAGCGUGACCACU .(((((..(..---....((..((((((((((((((......(((....))).---------....))))))))))))))..))(-(((((....)))))).........)..))))). ( -39.30) >DroAna_CAF1 73848 113 - 1 AGAGAAUUCUUUUGGAUGAGACUGG---UCUAGACCAGAAAUAGAUCAUUCUUGGCCCCCAUGUGAGGACUAUAACUCGAAAACGAUUCCCAAUGGGGGGCUAUUGUUA---GCCCACU ((((....)))).((..((((.(((---((((.........))))))).))))..))(((((.((.(((.......(((....))).))))))))))((((((....))---))))... ( -32.80) >consensus UGUGGAUCA_____ACUAAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUU_________UCGAGGGCCAUAACUUUAAGUUG_CUCCCCAUGGGGGGCUAUUGAAG_GUGACCACU .((((.................((((((((((((((......(((....)))..............))))))))))))))......(((((....)))))..............)))). (-23.09 = -23.90 + 0.81)

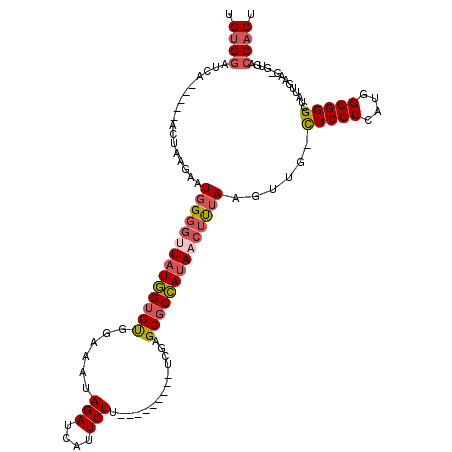
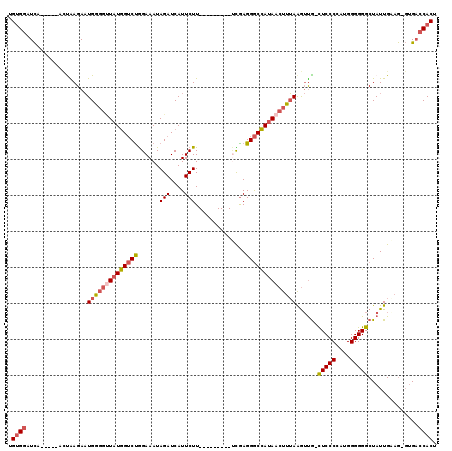
| Location | 12,218,105 – 12,218,211 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.09 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -20.64 |
| Energy contribution | -23.04 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12218105 106 - 20766785 UCUUCUAGAUCUAGGGCCUAGAUCGUUUUACAACUUCGCAUGUGGAUCA--ACUAAGAAUGGGGCUAUGGUCUGGAAAUAGAUCAUUCUUUCGAGGGCCAUAACUUUA ..((((.(((((((...)))))))(.((((((........)))))).).--....))))(((((.((((((((.(((..(((....))))))..)))))))).))))) ( -26.50) >DroSec_CAF1 79121 106 - 1 UCUUCUAGAUCUAGAGCCUAGAUCGUUUUACAACUUUGCAUGUGGAUCA--ACUAAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUUUCGAGGGCCAUAACUUUA ..((((.(((((((...)))))))(.((((((........)))))).).--....))))((((((((((((((.(((..(((....))))))..)))))))))))))) ( -30.70) >DroSim_CAF1 75552 106 - 1 UCUUCUAGAUCUAGAGCCUAGAUCGUUUUACAACUUCGCAUGUGGAUCA--ACUAAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUUUCGAGGGCCAUAACUUUA ..((((.(((((((...)))))))(.((((((........)))))).).--....))))((((((((((((((.(((..(((....))))))..)))))))))))))) ( -30.70) >DroEre_CAF1 87170 103 - 1 AUUUCAA-----AGAGCCUAGAUUAUUUUACAGCUUCGCAUGUGGAUCCAGAUCGAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUUUUAAGGGCCAUAACUUUA ..(((..-----..(((.((((....))))..)))(((.((.((....)).))))))))((((((((((((((..(((.(((....))))))..)))))))))))))) ( -26.00) >DroYak_CAF1 82130 103 - 1 GUUUCAG-----AGAGGCUAGAUCUUUUUACAGCUUCGCAUGUGGUUCCAGAUCAAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUUCUGAGGGCCAUAACUUUA (((((..-----.)))))..(((((..(((((((...)).)))))....))))).....((((((((((((((......(((....))).....)))))))))))))) ( -27.70) >consensus UCUUCUAGAUCUAGAGCCUAGAUCGUUUUACAACUUCGCAUGUGGAUCA__ACUAAGAAUGGGGUUAUGGUCUGGAAAUAGAUCAUUCUUUCGAGGGCCAUAACUUUA ..(((..(((((((...)))))))..((((((........))))))..........)))((((((((((((((......(((....))).....)))))))))))))) (-20.64 = -23.04 + 2.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:31 2006