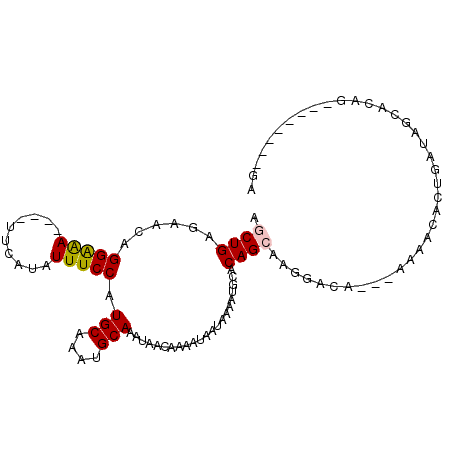
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,214,750 – 12,214,841 |
| Length | 91 |
| Max. P | 0.736545 |

| Location | 12,214,750 – 12,214,841 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -16.43 |
| Consensus MFE | -12.45 |
| Energy contribution | -12.32 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12214750 91 + 20766785 UC--------CUGUGCUAUCAGUGUUUU---UCUUCUUGCUGUGCAUUUUAUUAUUUUGUUAUUUGCAUUUGCAUGGAAAUAUGAA----UUCCCUGUUCUCAGCU ..--------....(((.........((---(((...(((.(((((....((......))....)))))..))).)))))...(((----(.....))))..))). ( -14.60) >DroPse_CAF1 78018 96 + 1 CA--------CU--CCUACAGCUGCUACUCCCGUCCUUGCUGUGCAUUUUAUUAUUUUGUUAUUUGCAUUUGCAUGGAAAUAUGAAAUAUUCUCCUGUUCUCAGUC .(--------((--...((((..(....((...(((.(((.(((((....((......))....)))))..))).))).....))......)..))))....))). ( -13.80) >DroEre_CAF1 83740 99 + 1 UCCUGUAGUCCUGUGCUAUCAGUGUUUU---CCUCCUUGCUGUGCAUUUUAUUAUUUUGUUAUUUGCAUUUGCAUGGAAAUAUGAA----UUCCCUGUUCUCAGCU ..(((.((..(.......(((....(((---((....(((.(((((....((......))....)))))..))).)))))..))).----......)..))))).. ( -17.24) >DroYak_CAF1 78825 91 + 1 UG--------CUGUGCUAUCAGUGUUUU---UCUCCAUGCUGUGCAUUUUAUUAUUUUGUUAUUUGCAUUUGCAUGGAAAUAUGAA----UUCCCUGUUCUCAGCU .(--------(((.(....(((.((((.---..(((((((.(((((....((......))....)))))..))))))).....)))----)...)))..).)))). ( -23.80) >DroAna_CAF1 70914 95 + 1 CC--------UUACCCCAUACGUGCUUU-UGUGUCCUUGCUGUGCAUUUUAUUAUUUCGUUAUUUGCAUUUGCAUGGAAAUAUGAAA--CUCUCCUGUUCUCAGCU ..--------.............((...-(((.(((.(((.(((((..................)))))..))).))).)))(((((--(......))).))))). ( -15.37) >DroPer_CAF1 78669 96 + 1 CA--------CU--CCUACAGCUGCUACUCCCGUCCUUGCUGUGCAUUUUAUUAUUUUGUUAUUUGCAUUUGCAUGGAAAUAUGAAAUAUUCUCCUGUUCUCAGUC .(--------((--...((((..(....((...(((.(((.(((((....((......))....)))))..))).))).....))......)..))))....))). ( -13.80) >consensus CC________CUGUCCUAUAAGUGCUUU___CCUCCUUGCUGUGCAUUUUAUUAUUUUGUUAUUUGCAUUUGCAUGGAAAUAUGAA____UCCCCUGUUCUCAGCU .................................(((.(((.(((((....((......))....)))))..))).)))................(((....))).. (-12.45 = -12.32 + -0.14)

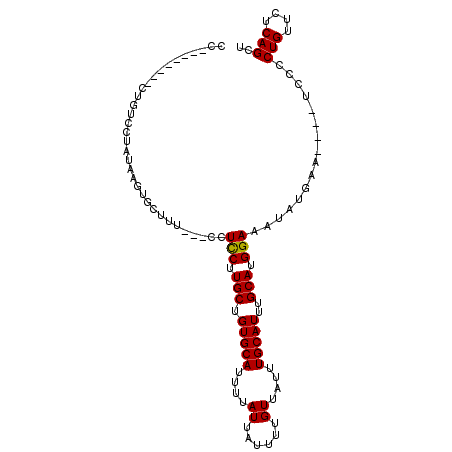
| Location | 12,214,750 – 12,214,841 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -16.83 |
| Consensus MFE | -10.02 |
| Energy contribution | -9.85 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12214750 91 - 20766785 AGCUGAGAACAGGGAA----UUCAUAUUUCCAUGCAAAUGCAAAUAACAAAAUAAUAAAAUGCACAGCAAGAAGA---AAAACACUGAUAGCACAG--------GA .((((......(((((----(....)))))).(((....)))......................)))).......---......(((......)))--------.. ( -15.70) >DroPse_CAF1 78018 96 - 1 GACUGAGAACAGGAGAAUAUUUCAUAUUUCCAUGCAAAUGCAAAUAACAAAAUAAUAAAAUGCACAGCAAGGACGGGAGUAGCAGCUGUAGG--AG--------UG ..(((....)))...............((((.(((....))).....................(((((....((....))....))))).))--))--------.. ( -16.40) >DroEre_CAF1 83740 99 - 1 AGCUGAGAACAGGGAA----UUCAUAUUUCCAUGCAAAUGCAAAUAACAAAAUAAUAAAAUGCACAGCAAGGAGG---AAAACACUGAUAGCACAGGACUACAGGA .((((......(((((----(....)))))).(((....)))......................)))).......---......(((.((((....).)))))).. ( -17.20) >DroYak_CAF1 78825 91 - 1 AGCUGAGAACAGGGAA----UUCAUAUUUCCAUGCAAAUGCAAAUAACAAAAUAAUAAAAUGCACAGCAUGGAGA---AAAACACUGAUAGCACAG--------CA .((((.(..(((.(..----..)...(((((((((...((((..................))))..)))))))))---......)))....).)))--------). ( -21.47) >DroAna_CAF1 70914 95 - 1 AGCUGAGAACAGGAGAG--UUUCAUAUUUCCAUGCAAAUGCAAAUAACGAAAUAAUAAAAUGCACAGCAAGGACACA-AAAGCACGUAUGGGGUAA--------GG .((((......(((((.--.......))))).(((....)))......................)))).........-..................--------.. ( -13.80) >DroPer_CAF1 78669 96 - 1 GACUGAGAACAGGAGAAUAUUUCAUAUUUCCAUGCAAAUGCAAAUAACAAAAUAAUAAAAUGCACAGCAAGGACGGGAGUAGCAGCUGUAGG--AG--------UG ..(((....)))...............((((.(((....))).....................(((((....((....))....))))).))--))--------.. ( -16.40) >consensus AGCUGAGAACAGGAAA____UUCAUAUUUCCAUGCAAAUGCAAAUAACAAAAUAAUAAAAUGCACAGCAAGGACA___AAAACACUGAUAGCACAG________GA .((((......(((((..........))))).(((....)))......................))))...................................... (-10.02 = -9.85 + -0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:27 2006