| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,213,395 – 12,213,497 |
| Length | 102 |
| Max. P | 0.937133 |

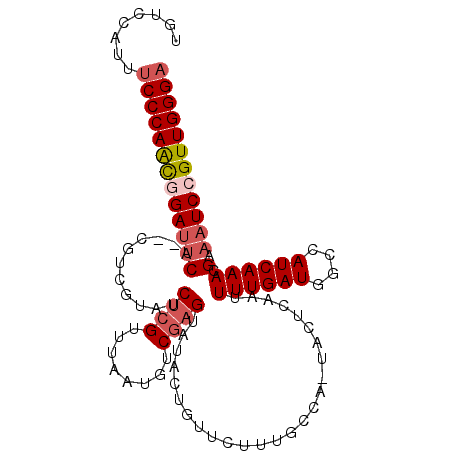
| Location | 12,213,395 – 12,213,497 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -23.29 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12213395 102 + 20766785 UCCCAACGGAUUUCCUUUGAUGGCCAUCAAAAUUGAGUA-UGGCAAAGAACAGUAUACUCGUCAUUAAACGAGAUACGACG--UGAAUCCGUUGGGAAAUGGACA ((((((((((((..(.......((((((........).)-))))........((((.(((((......)))))))))...)--..))))))))))))........ ( -37.50) >DroSec_CAF1 74485 103 + 1 UCCCAACGGAUUUCCUUUGAUGGCCAUCAAAAUUGAGCAAAGGCAAAGAGCAGUAUACUCGACAUUAACCGAGAAACGACG--UGAAUCAGUUGGGAAAUGGACA (((((((.((((..(.(((...(((.(((....))).....))).............((((........))))...))).)--..)))).)))))))........ ( -26.40) >DroSim_CAF1 70864 103 + 1 UCCCAACGGAUUUCCUUUGAUGGCCAUCAAACUUGAGCAAAGGCAAAGAGCAGUAUACUCGACAUUAAACGAGAAACGACG--UGAAUCCGUUGGGAAAUGGACA ((((((((((((..(.(((...(((.(((....))).....))).............((((........))))...))).)--..))))))))))))........ ( -32.10) >DroEre_CAF1 82397 102 + 1 UCCCAACGGAUUUCGUUUGAUGGCCAUCAAAAUUGAGUU-UGGCAAUGAAGAGAAUACUCGACAUUCAACGAGAAACGACG--UGAAUCUGCUGGGAAAUGGACG (((((.((((((((((((..(((((((((....)))...-))))((((..(((....)))..))))...))..))))))..--..)))))).)))))........ ( -28.60) >DroYak_CAF1 77476 103 + 1 UCCCAACGGAUUUCGUUUGAUGGCCAUCAAAAUUGAGUU-UGGCAAAGAAAAGAAUACUCGACAUUCAACGAGAUACGACA-ACGAAUCCGCUGGGAAAUGGACU (((((.((((((.((((.(.(((((((((....)))...-)))).............((((........))))...)).))-))))))))).)))))........ ( -29.80) >DroAna_CAF1 69511 104 + 1 ACCCAACGGAUUUCGUUUGAUGGCCAUCAAAAUUGAGUA-UCACAUACAACGUUAGACAUGGUCAUAAACGAGAUUCCAUCGGAGCGUCCAUUGGGGAAAAGCAA .(((((.((((((((((((.(((((((((....)))(((-(...))))...........))))))))))))))))))....((.....)).)))))......... ( -30.60) >consensus UCCCAACGGAUUUCCUUUGAUGGCCAUCAAAAUUGAGUA_UGGCAAAGAACAGUAUACUCGACAUUAAACGAGAAACGACG__UGAAUCCGUUGGGAAAUGGACA (((((((((((((..((((((....))))))..........................((((........))))...........)))))))))))))........ (-23.29 = -24.07 + 0.78)

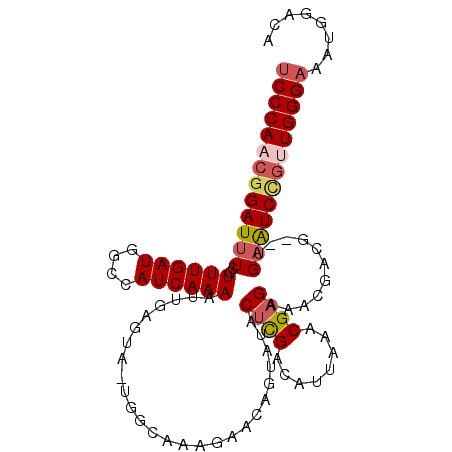
| Location | 12,213,395 – 12,213,497 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -21.23 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12213395 102 - 20766785 UGUCCAUUUCCCAACGGAUUCA--CGUCGUAUCUCGUUUAAUGACGAGUAUACUGUUCUUUGCCA-UACUCAAUUUUGAUGGCCAUCAAAGGAAAUCCGUUGGGA ........((((((((((((..--(...((((((((((....)))))).))))........((((-(.(........)))))).......)..)))))))))))) ( -37.60) >DroSec_CAF1 74485 103 - 1 UGUCCAUUUCCCAACUGAUUCA--CGUCGUUUCUCGGUUAAUGUCGAGUAUACUGCUCUUUGCCUUUGCUCAAUUUUGAUGGCCAUCAAAGGAAAUCCGUUGGGA ........(((((((.((((..--((.((((........)))).)(((((....((.....))...)))))...((((((....)))))))..)))).))))))) ( -27.30) >DroSim_CAF1 70864 103 - 1 UGUCCAUUUCCCAACGGAUUCA--CGUCGUUUCUCGUUUAAUGUCGAGUAUACUGCUCUUUGCCUUUGCUCAAGUUUGAUGGCCAUCAAAGGAAAUCCGUUGGGA ........((((((((((((..--((.((((........)))).)(((((....((.....))...)))))...((((((....)))))))..)))))))))))) ( -33.40) >DroEre_CAF1 82397 102 - 1 CGUCCAUUUCCCAGCAGAUUCA--CGUCGUUUCUCGUUGAAUGUCGAGUAUUCUCUUCAUUGCCA-AACUCAAUUUUGAUGGCCAUCAAACGAAAUCCGUUGGGA ........(((((((.((((..--..((((((.......((((..(((....)))..))))((((-...(((....)))))))....)))))))))).))))))) ( -26.70) >DroYak_CAF1 77476 103 - 1 AGUCCAUUUCCCAGCGGAUUCGU-UGUCGUAUCUCGUUGAAUGUCGAGUAUUCUUUUCUUUGCCA-AACUCAAUUUUGAUGGCCAUCAAACGAAAUCCGUUGGGA ........(((((((((((((((-(...(((.((((........)))))))..........((((-...(((....))))))).....)))).)))))))))))) ( -32.40) >DroAna_CAF1 69511 104 - 1 UUGCUUUUCCCCAAUGGACGCUCCGAUGGAAUCUCGUUUAUGACCAUGUCUAACGUUGUAUGUGA-UACUCAAUUUUGAUGGCCAUCAAACGAAAUCCGUUGGGU .........(((((((((...(((...)))...(((((((((.((((...(((.((((...(...-..).)))).))))))).))).))))))..))))))))). ( -26.50) >consensus UGUCCAUUUCCCAACGGAUUCA__CGUCGUAUCUCGUUUAAUGUCGAGUAUACUGUUCUUUGCCA_UACUCAAUUUUGAUGGCCAUCAAACGAAAUCCGUUGGGA ........(((((((((((((...........((((........))))..........................((((((....)))))).).)))))))))))) (-21.23 = -22.03 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:25 2006