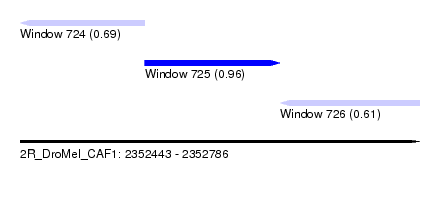
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,352,443 – 2,352,786 |
| Length | 343 |
| Max. P | 0.955623 |

| Location | 2,352,443 – 2,352,550 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2352443 107 - 20766785 AUCCGCAAAAGCCGCAAAACUUUGGCUAGCCGCCAUCAUUAGCCGUUGUCAGGCGUC-------------AGGCUGUCAUUAAAAAUGGCUUAAACAUCAGCCCAGUAGGCGGUUGAAAA .........(((((........)))))(((((((.......(((.......)))...-------------.(((((...((((.......))))....))))).....)))))))..... ( -30.30) >DroSec_CAF1 2050 107 - 1 AUCCGCAAAAGCCGCAAAACUUUGGCUAGCCGCCAUCAUUAGCCGUUGUCAGGCGUC-------------AGGCUGUCAUUAAAAAUGGCUUAAACAUCAGCCCAGUAGGCGGUCGAAAA .........(((((........))))).((((((.......(((.......)))...-------------.(((((...((((.......))))....))))).....))))))...... ( -29.20) >DroSim_CAF1 2074 107 - 1 AUCCGCAAAAGCCGCAAAACUUUGGCUAGCCGCCAUCAUUAGCCGUUGUCAGGCGUC-------------AGGCUGUCAUUAAAAAUGGCUUAAACAUCAGCCCAGUAGGCGGUCGAAAA .........(((((........))))).((((((.......(((.......)))...-------------.(((((...((((.......))))....))))).....))))))...... ( -29.20) >DroEre_CAF1 1912 120 - 1 AUCCGCAAAAGCCGCAAAACUUUGGCUAGCCGCCAUCAUUAGCCGUUGUCAGGCGCCAGGCGCCAGCGCCAGGCUGUCAUUAAAAAUGGCUUAAACAUCAGGCCAGCAGGCGGUUGAAAA .........(((((........)))))(((((((.......(((.......)))(((.((((....)))).)))............((((((.......))))))...)))))))..... ( -42.50) >DroYak_CAF1 2048 107 - 1 AUCCGCAAAAGCCGCAAAACUUUGGCUAGCCGCCAUCAUUAGCCGUUGUCAGGCGUC-------------AGUUUGUCAUUAAAAAUGGCUUAAACAUCAGGCCAGCAGGCGGUUGAUAC .........(((((........)))))(((((((............((.(((((...-------------.))))).)).......((((((.......))))))...)))))))..... ( -30.40) >consensus AUCCGCAAAAGCCGCAAAACUUUGGCUAGCCGCCAUCAUUAGCCGUUGUCAGGCGUC_____________AGGCUGUCAUUAAAAAUGGCUUAAACAUCAGCCCAGUAGGCGGUUGAAAA .........(((((........)))))(((((((.......(((.......))).................(((((...((((.......))))....))))).....)))))))..... (-27.12 = -27.32 + 0.20)

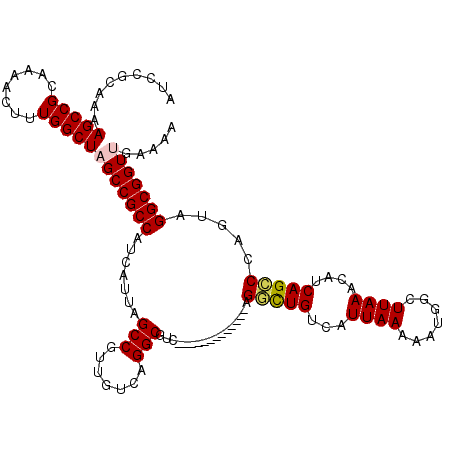
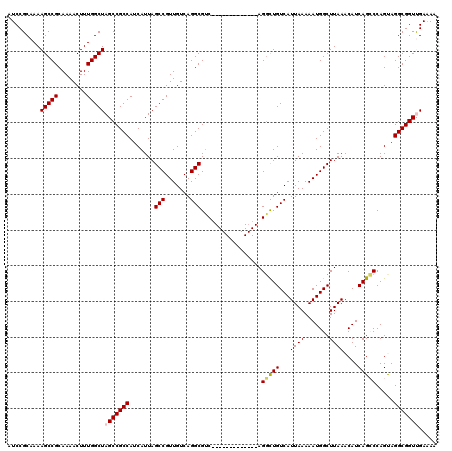
| Location | 2,352,550 – 2,352,666 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2352550 116 + 20766785 UAAAAUUGAAAACGCUUGCAUUCAUUAAUCCUGCCAACGGAAA----AACCCAACUAGAACGUGACAUGAGGCAGGUGUCGCAUUAAUUAGUGUAAGCAUUCACACGGCUCCCACUCCAC ......((((...(((((((((.((((((((((((...((...----...)).((......)).......))))))......)))))).))))))))).))))...((........)).. ( -26.70) >DroSec_CAF1 2157 116 + 1 UAAAAUUGAAAACGCUUGCAUUCAUUAAUCCCGCCAACUGAAA----AACCCAACUAGAACGUGACAUGAGGCAGGUGUCGCAUUAAUUAGUGUAAGCAUUCACACGGCUCCCACUCCAC ......((((...(((((((((.((((((........(((...----........)))...(((((((.......))))))))))))).))))))))).))))...((........)).. ( -22.30) >DroSim_CAF1 2181 116 + 1 UAAAAUUGAAAACGCUUGCAUUCAUUAAUCCCGCCAACGGAAA----AACCCAACUAGAACGUGACAUGAGGCAGGUGUCGCAUUAAUUAGUGUAAGCAUUCACACGGCUCCCACUCCAC ......((((...(((((((((.((((((.(((....)))...----..............(((((((.......))))))))))))).))))))))).))))...((........)).. ( -26.30) >DroEre_CAF1 2032 115 + 1 UAAAAUUGAAAACGC-UGCAUUAAUUAAUCCCGCCAACGGAAG----AAUCCAACUAAAACGUGACAUGAGACAGGUGUCGCAUUAAUUAGUGCAAGCAUUCACACGGCUCCCUCUCCAC ......((((...((-(((((((((((((.(((....)))...----..............(((((((.......))))))))))))))))))).))).))))...((........)).. ( -27.50) >DroYak_CAF1 2155 120 + 1 UAAAAUUGAAAACGCUUGCAUUCAUUAAUCCCGCCAACAGAAAAAAAAACCCAACUAGAACGUGACAUGAGACAGGUGUCGCAUUAAUUAGUGUAAGCAUUCAUACGGCUCCCACUCCAC ......((((...(((((((((.((((((..(.......).....................(((((((.......))))))))))))).))))))))).))))...((........)).. ( -21.50) >consensus UAAAAUUGAAAACGCUUGCAUUCAUUAAUCCCGCCAACGGAAA____AACCCAACUAGAACGUGACAUGAGGCAGGUGUCGCAUUAAUUAGUGUAAGCAUUCACACGGCUCCCACUCCAC ......((((...(((((((((.((((((.(((....))).....................(((((((.......))))))))))))).))))))))).))))...((........)).. (-23.04 = -23.40 + 0.36)

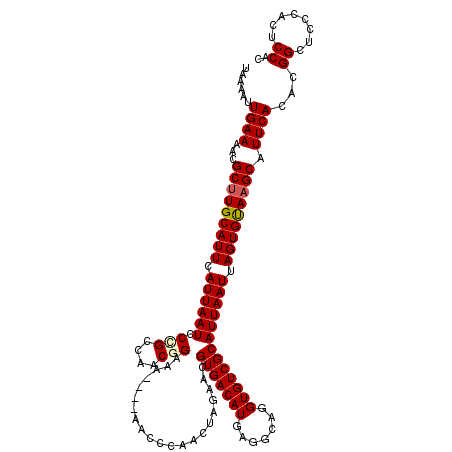
| Location | 2,352,666 – 2,352,786 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.56 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2352666 120 - 20766785 AAGACGGUGCGGAAUGCCGUUUGCAAUAAUUUAACUCAAGCCAUCUUGACCCAAAUUGCGGAGUGACACAUCUCGGCGAAAUGAAACCGCCCGCCCGAGGAAACUUUCAUUUCGCAAGCU .(((((((.......)))))))((...........(((((....)))))......(((((((((((........((((.........)))).....(((....)))))))))))))))). ( -34.70) >DroSec_CAF1 2273 120 - 1 AAGACGGUGCGGAAUGCCGUUUGCAAUAAUUUAACUCAAGCCAUCUUGACCCAAAUUGCGGAGUGACACAUCUCGGCGAAAUGAAACCGCCCGCCCGAGGAAACUUUCAUUUCGCAAGCU .(((((((.......)))))))((...........(((((....)))))......(((((((((((........((((.........)))).....(((....)))))))))))))))). ( -34.70) >DroSim_CAF1 2297 120 - 1 AAGACGGUGCGGAAUGCCGUUUGCAAUAAUUUAACUCAAGCCAUCUUGACCCAAAUUGCGGAGUGACACAUCUCGGCGAAAUGAAACCGCCCGCCCGAGGAAACUUUCAUUUCGCAAGCU .(((((((.......)))))))((...........(((((....)))))......(((((((((((........((((.........)))).....(((....)))))))))))))))). ( -34.70) >DroEre_CAF1 2147 120 - 1 AAGUCGGUGCUGAACGGCGCUUGCAAUAAUUUAACUCAAGCCAUCUUGACCCAAAUUGCGGAGUGACACAUCUCGGUGAAAUUAAAACGCCCGCCCGAGGAAACUUUCUUUUCGCAAGAU ..((.((((((....)))))).))...........(((((....)))))......((((((((.((........((((.............)))).(((....))))).))))))))... ( -28.12) >DroYak_CAF1 2275 120 - 1 AAGACGGUGCGGAAUGCCGUUUGCAAUAAUUUAACUCAAGCCAUCUUGACCCAAAUUGCGAAGUGACACAUCUCGGCGAAAUUAAACCGCCCGCCCGAGGAAACUUUCAUUUUGCAAGAC .(((((((.......))))))).............(((((....)))))......(((((((((((........((((.........)))).....(((....))))))))))))))... ( -31.20) >consensus AAGACGGUGCGGAAUGCCGUUUGCAAUAAUUUAACUCAAGCCAUCUUGACCCAAAUUGCGGAGUGACACAUCUCGGCGAAAUGAAACCGCCCGCCCGAGGAAACUUUCAUUUCGCAAGCU .(((((((.......))))))).............(((((....)))))......(((((((((((........((((.........)))).....(((....))))))))))))))... (-29.44 = -29.56 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:37 2006