| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,203,014 – 12,203,104 |
| Length | 90 |
| Max. P | 0.989389 |

| Location | 12,203,014 – 12,203,104 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -19.66 |
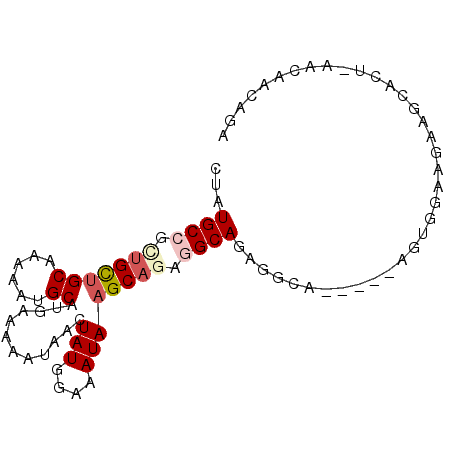
| Consensus MFE | -7.50 |
| Energy contribution | -8.06 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12203014 90 + 20766785 UCAGUUGUU-AGUGCUUCUUCCACUUCCUGUGCCUCUGCCUCUGCUUAUUUCCAUAGUUAUUUUUCAUGCAUUUUUGCAGCAACGGCAUAG .........-((((.......))))..(((((((..(((....(((.........))).........((((....)))))))..))))))) ( -17.10) >DroSim_CAF1 58754 90 + 1 UAAAUUGAU-AGAGCCUCUUUCACUUCAUGUGCCUCUGCCUUUGCUUAUUUCUAUACUUAUUUGUCAUGCAUUUAAGCAGCAGCGGCAUAA .....(((.-((.....)).))).....((((((.((((...((((((..(.(((((......)).))).)..)))))))))).)))))). ( -23.40) >DroEre_CAF1 72165 81 + 1 UCUG---UU-AGUGCUUCUGCCACA------GCCACUGCCUCUGCUUAUUUCCAUAGUUAUUUUUCAUGCAUUUUUGCUGCAGCGGCAUAG .(((---(.-.(.((....))))))------)....((((.((((...........(........)..(((....))).)))).))))... ( -16.10) >DroYak_CAF1 67105 84 + 1 UCUGUUGUU-AGUGCUUCUGCCAGU------UCCUCUGCGUCUGCUUAUUUCCAUAGUUAUUUUUCAUGCAUUUUUGCAGCAGCGGCAUAG .........-.(((((.(((((((.------....))).............................((((....)))))))).))))).. ( -16.90) >DroAna_CAF1 59383 76 + 1 UCUGUUGUGUUAUGGAUC------------UGGCUCUGC---UGCUUAUUUCCAUAGUUAUUUUUCAUGCAUUUUUGCAACGGCAGCAGAG ((((((((.(((((((..------------.(((.....---.)))....)))))))..........((((....))))...)))))))). ( -24.80) >consensus UCUGUUGUU_AGUGCUUCUGCCACU_____UGCCUCUGCCUCUGCUUAUUUCCAUAGUUAUUUUUCAUGCAUUUUUGCAGCAGCGGCAUAG ...............................(((.((((...(((.......(((((......)).))).......))))))).))).... ( -7.50 = -8.06 + 0.56)

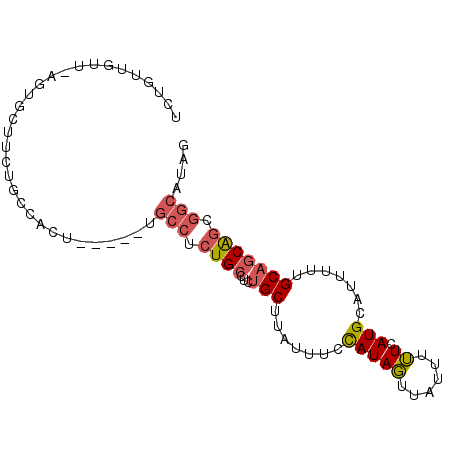
| Location | 12,203,014 – 12,203,104 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -19.44 |
| Consensus MFE | -10.26 |
| Energy contribution | -11.22 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12203014 90 - 20766785 CUAUGCCGUUGCUGCAAAAAUGCAUGAAAAAUAACUAUGGAAAUAAGCAGAGGCAGAGGCACAGGAAGUGGAAGAAGCACU-AACAACUGA ((.((((.((((((((....)))............(((....)))))))).)))).))...(((..((((.......))))-.....))). ( -19.90) >DroSim_CAF1 58754 90 - 1 UUAUGCCGCUGCUGCUUAAAUGCAUGACAAAUAAGUAUAGAAAUAAGCAAAGGCAGAGGCACAUGAAGUGAAAGAGGCUCU-AUCAAUUUA ...((((.(((((((......))...........((.((....)).))...))))).))))..((((.(((.((.....))-.))).)))) ( -20.70) >DroEre_CAF1 72165 81 - 1 CUAUGCCGCUGCAGCAAAAAUGCAUGAAAAAUAACUAUGGAAAUAAGCAGAGGCAGUGGC------UGUGGCAGAAGCACU-AA---CAGA (((((((.((((.(((....)))............(((....))).)))).)))).)))(------(((.((....))...-.)---))). ( -22.00) >DroYak_CAF1 67105 84 - 1 CUAUGCCGCUGCUGCAAAAAUGCAUGAAAAAUAACUAUGGAAAUAAGCAGACGCAGAGGA------ACUGGCAGAAGCACU-AACAACAGA ((.(((..((((((((....)))............(((....))))))))..))).))..------.(((...........-.....))). ( -15.29) >DroAna_CAF1 59383 76 - 1 CUCUGCUGCCGUUGCAAAAAUGCAUGAAAAAUAACUAUGGAAAUAAGCA---GCAGAGCCA------------GAUCCAUAACACAACAGA ((((((((((((.(((....)))))).........(((....))).)))---))))))...------------.................. ( -19.30) >consensus CUAUGCCGCUGCUGCAAAAAUGCAUGAAAAAUAACUAUGGAAAUAAGCAGAGGCAGAGGCA_____AGUGGAAGAAGCACU_AACAACAGA ...((((.(((((((......))............(((....)))))))).)))).................................... (-10.26 = -11.22 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:23 2006