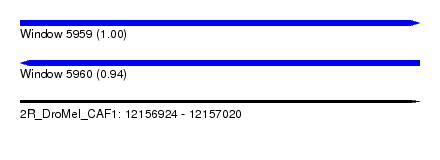
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,156,924 – 12,157,020 |
| Length | 96 |
| Max. P | 0.999034 |

| Location | 12,156,924 – 12,157,020 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 71.12 |
| Mean single sequence MFE | -15.12 |
| Consensus MFE | -7.97 |
| Energy contribution | -9.72 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12156924 96 + 20766785 UUAAUAUUAAGCUAGCAAACGGAUUUAACUAAUCCGUUGCUGAUAACAAAAACUUUAUGUGUAAAAUUCAACUUACAAAAAAAAAAACAAUAUCUG ...(((((....((((((.((((((.....)))))))))))).................(((((........)))))...........)))))... ( -15.70) >DroSec_CAF1 12892 78 + 1 UUAAUAUUAAGCUAGCAAACGGAUUUAACUAAUCCGUUGCUAACAACAAAAACUGUAUGUGUCAAAUUCA--------AAAAUUUA---------- ............((((((.((((((.....))))))))))))((((((.....))).)))..........--------........---------- ( -15.00) >DroSim_CAF1 12993 86 + 1 UUAAUAUUAAGCUAGCAAACGGAUUUAACUAAUCCGUUGCUAACAACAAUAACUGUAUGUGUAAAAUUCAACUUACAAAAAAUUUA---------- ..........(.((((((.((((((.....)))))))))))).).........((((.((..........)).)))).........---------- ( -16.30) >DroEre_CAF1 18452 80 + 1 UUAAUAUUUAGCUAGCAUAGCUAGCUA-----------GCUGAUAACAAAAC-UGUAUGUGUAACAUUCAACUUUAA--AAAUUUU--AAUAUCUG .......(((((((((.......))))-----------))))).........-((.(((.....))).)).......--.......--........ ( -13.50) >consensus UUAAUAUUAAGCUAGCAAACGGAUUUAACUAAUCCGUUGCUAACAACAAAAACUGUAUGUGUAAAAUUCAACUUACA_AAAAUUUA__________ ............((((((.((((((.....))))))))))))...................................................... ( -7.97 = -9.72 + 1.75)


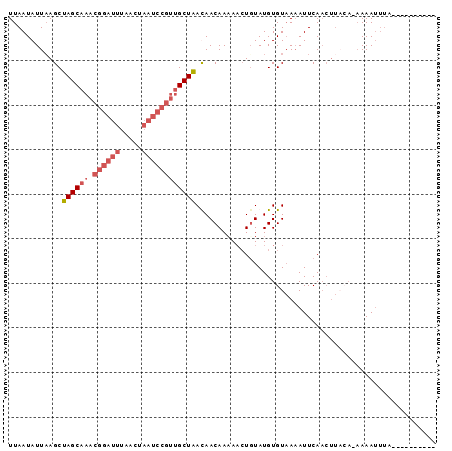
| Location | 12,156,924 – 12,157,020 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 71.12 |
| Mean single sequence MFE | -18.66 |
| Consensus MFE | -7.50 |
| Energy contribution | -10.50 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12156924 96 - 20766785 CAGAUAUUGUUUUUUUUUUUGUAAGUUGAAUUUUACACAUAAAGUUUUUGUUAUCAGCAACGGAUUAGUUAAAUCCGUUUGCUAGCUUAAUAUUAA ..((((.......((((..((((((......))))))...)))).......))))(((((((((((.....)))))).)))))............. ( -18.44) >DroSec_CAF1 12892 78 - 1 ----------UAAAUUUU--------UGAAUUUGACACAUACAGUUUUUGUUGUUAGCAACGGAUUAGUUAAAUCCGUUUGCUAGCUUAAUAUUAA ----------........--------..........................((((((((((((((.....)))))).)))))))).......... ( -17.50) >DroSim_CAF1 12993 86 - 1 ----------UAAAUUUUUUGUAAGUUGAAUUUUACACAUACAGUUAUUGUUGUUAGCAACGGAUUAGUUAAAUCCGUUUGCUAGCUUAAUAUUAA ----------.........((((((......))))))........(((((..((((((((((((((.....)))))).)))))))).))))).... ( -21.90) >DroEre_CAF1 18452 80 - 1 CAGAUAUU--AAAAUUU--UUAAAGUUGAAUGUUACACAUACA-GUUUUGUUAUCAGC-----------UAGCUAGCUAUGCUAGCUAAAUAUUAA ..((((((--.......--.((((..((.(((.....))).))-..)))).....(((-----------((((.......))))))).)))))).. ( -16.80) >consensus __________UAAAUUUU_UGUAAGUUGAAUUUUACACAUACAGUUUUUGUUAUCAGCAACGGAUUAGUUAAAUCCGUUUGCUAGCUUAAUAUUAA ....................................................((((((((((((((.....)))))).)))))))).......... ( -7.50 = -10.50 + 3.00)

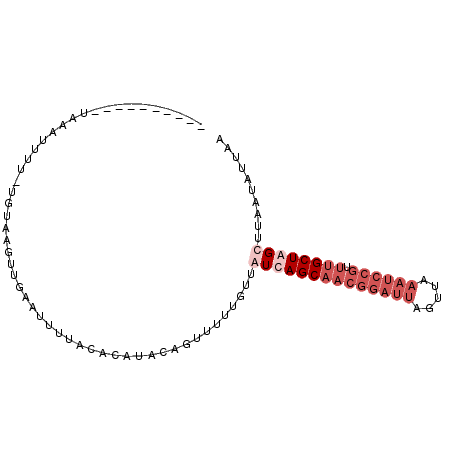

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:13 2006