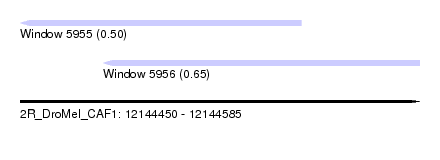
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,144,450 – 12,144,585 |
| Length | 135 |
| Max. P | 0.652790 |

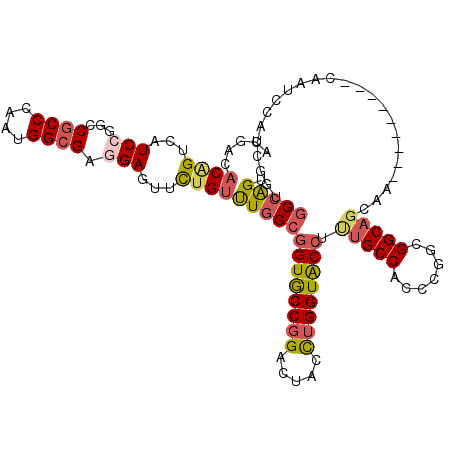
| Location | 12,144,450 – 12,144,545 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -29.75 |
| Energy contribution | -29.50 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12144450 95 - 20766785 UUUGGCGGUGCCGGAUUAUCUGGUACCUUUGCCACCCGGCGGCAGCAA---------CAAUCCAUCGAUGGCCAGUGGUUCG---GGCUACGUGCCAGAAUUGCAAC ..((((((((((((.....)))))))).(((((.......)))))...---------.............))))(..((((.---(((.....))).))))..)... ( -38.60) >DroSec_CAF1 516 95 - 1 UUUGGCGGUGCCGGAUUACCUGGUACCUUUGCCACCCGGUGGCAGCAA---------CAAUCCAUCAGUGGCCAGUGGUUCG---GGCUACGUGCCAGAAUUGCAAC ..((((((((((((.....)))))))).(((((((...)))))))...---------.............))))(..((((.---(((.....))).))))..)... ( -41.00) >DroSim_CAF1 516 95 - 1 UUUGGCGGUGCCGGAUUACCUGGUACCUUUGCCACCCGGCGGCAGUAA---------CAAUCCAUCAGUGGCCAGUGGUUCG---GGCUACGUGCCAGAAUUGCAAC ..((((((((((((.....)))))))).(((((.......)))))...---------.............))))(..((((.---(((.....))).))))..)... ( -38.10) >DroEre_CAF1 6115 95 - 1 UUUGGCGGUGCCGGACUACCUGGUACCUUUGCCACCCGGCGGCAGCAA---------CAAUCCAUCGGUGGCCAGUGGUUCG---GGCUACGUCCCGGAAUCGCAAC ..((((((((((((.....)))))))).....((((.(..((......---------....))..)))))))))((((((((---((......))).)))))))... ( -37.60) >DroYak_CAF1 6128 95 - 1 CUUGGCGGUACCUGACUACCUGGUACCUUUGCCACCCGGCGGCAGCAA---------CAAUCCAUCGGUGGCCAGCGGAUCG---GGCUACGUCCCAGAAUUGCAAC ..((((((((((.........)))))).....((((.(..((......---------....))..)))))))))((((.(((---((......))).)).))))... ( -29.70) >DroAna_CAF1 540 107 - 1 UCUGGCGGUGCCGGACUACUUGGUGCCCCUGCCCCCGGGUGGCAGCAGUGGCCUCGGCACCCCACAAAUGGCCAGUGGCUCCGGUGGCUACGGUGCGGAAUCGCAGC ..(((.((((((((.(((((.(.((((((((....)))).)))).)))))).).))))))))))....((((((.((....)).)))))).(.((((....)))).) ( -53.80) >consensus UUUGGCGGUGCCGGACUACCUGGUACCUUUGCCACCCGGCGGCAGCAA_________CAAUCCAUCAGUGGCCAGUGGUUCG___GGCUACGUGCCAGAAUUGCAAC ..((((((((((((.....)))))))).(((((.......))))).........................))))(((((((....((.......)).)))))))... (-29.75 = -29.50 + -0.25)

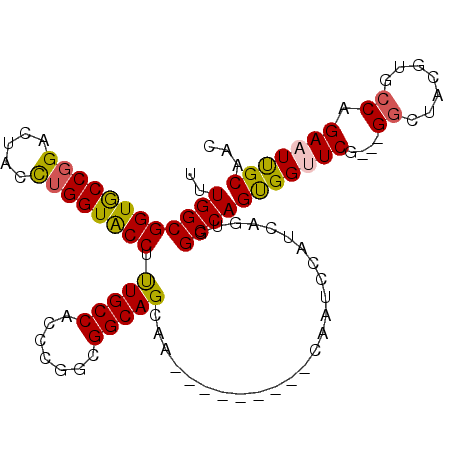
| Location | 12,144,478 – 12,144,585 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -44.12 |
| Consensus MFE | -30.85 |
| Energy contribution | -31.47 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12144478 107 - 20766785 ACGACACAGUCAUCCGGCCGCCCAAUGGCGAGGAGUUCUGUUUGGCGGUGCCGGAUUAUCUGGUACCUUUGCCACCCGGCGGCAGCAA---------CAAUCCAUCGAUGGCCAGU ..(((...)))....(((((((....)))(((((((((((((....((((((((.....))))))))...(((....)))))))).))---------)..))).))...))))... ( -41.30) >DroSec_CAF1 544 107 - 1 ACGACACAGUCAUCCGGCCGCCCAAUGGCGAAGAGUUCUGUUUGGCGGUGCCGGAUUACCUGGUACCUUUGCCACCCGGUGGCAGCAA---------CAAUCCAUCAGUGGCCAGU ..(((...)))....((((((...((((((((.(....).))))((((((((((.....))))))))..((((((...))))))))..---------....))))..))))))... ( -40.80) >DroEre_CAF1 6143 107 - 1 ACGACACAGUCAUCCGGCCGCCCAAUGGCGAGGAGUUCUGUUUGGCGGUGCCGGACUACCUGGUACCUUUGCCACCCGGCGGCAGCAA---------CAAUCCAUCGGUGGCCAGU ..(((...)))....(((((((..((((...(..((((((((((((((((((((.....))))))))...))))...))))).)))..---------)...)))).)))))))... ( -43.20) >DroYak_CAF1 6156 107 - 1 ACGACACAGUCAUCCGGCCGCCCAAUGGAGAGGAGUUCUGCUUGGCGGUACCUGACUACCUGGUACCUUUGCCACCCGGCGGCAGCAA---------CAAUCCAUCGGUGGCCAGC ..(((...)))....(((((((..(((((..(..((((((((((((((((((.........))))))...))))...))))).)))..---------)..))))).)))))))... ( -45.50) >DroMoj_CAF1 688 101 - 1 ACGAGACGGUCAUCCGGCCGCCCAAUGGCGAAGAGUUUUGUCUAGCCGUGCCGGACUAUCUGGUGCCGCUGCCUGGCAACGGC---------------GCGCCCCAUGCGCCCGGA (((((((((((....))))(((....))).....))))))).((((.(..((((.....))))..).))))((.(....)(((---------------(((.....)))))).)). ( -41.40) >DroAna_CAF1 571 116 - 1 AUGACACCGUCAUCCGGCCGCCCAACGGCGAGGAGUUCUGUCUGGCGGUGCCGGACUACUUGGUGCCCCUGCCCCCGGGUGGCAGCAGUGGCCUCGGCACCCCACAAAUGGCCAGU (((((...)))))..(((((((....))).............(((.((((((((.(((((.(.((((((((....)))).)))).)))))).).)))))))))).....))))... ( -52.50) >consensus ACGACACAGUCAUCCGGCCGCCCAAUGGCGAGGAGUUCUGUUUGGCGGUGCCGGACUACCUGGUACCUUUGCCACCCGGCGGCAGCAA_________CAAUCCAUCGGUGGCCAGU .....((((...(((...((((....)))).)))...))))(((((((((((((.....)))))))).(((((.......))))).........................))))). (-30.85 = -31.47 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:09 2006