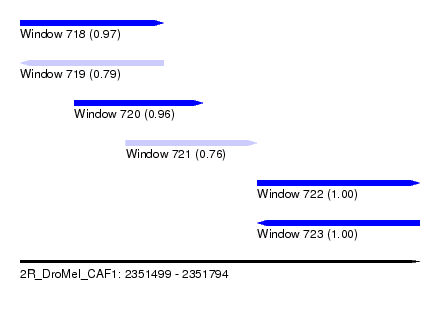
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,351,499 – 2,351,794 |
| Length | 295 |
| Max. P | 0.998699 |

| Location | 2,351,499 – 2,351,605 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.66 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2351499 106 + 20766785 AUGGCCAGCAGCAGUGAGGAGCCGAAGGUCUCAACUAAUUAAAUUUCCCAGAUGUGUGUGU--GUGUGUGUUUGCAUGCACAUAUUU------------AAAUUGGCUCUGCCAGGCCAC .(((((.((((.(((.(((((.(....).))).................((((((((((((--(((......)))))))))))))))------------...)).)))))))..))))). ( -38.30) >DroSec_CAF1 1105 108 + 1 AUGGCCAGCAGCAGUGAGGAGCCGAAGGUCUCAGCUAAUUAAAUUUCCCAGAUGUGUGUGUGUGUGUGUGUUUGCAUACACAUAUUU------------AAAUUGGCUCUGCCAGGCCAC .(((((.((((.(((.(((((.(....).))).................(((((((((((((((........)))))))))))))))------------...)).)))))))..))))). ( -39.10) >DroSim_CAF1 1120 102 + 1 AUGGCCAGCAGCAGUGAGGAGCCGAAGGUCUCAGCUAAUUAAAUUUCCCAGCUGUGUG------UGUGUGUUUGCAUACACAUAUUU------------AAAUUGGCUCUGCCAGGCCAC .(((((.((....))..((((((((......(((((.............)))))((((------((((((....))))))))))...------------...))))))))....))))). ( -37.42) >DroYak_CAF1 1120 116 + 1 AUGGCCAGCAGCAGGGACGAAACGAAGGUCUCAGCUAAUUAAAUUUACCAGAUGUGUAUG----UGGGUGUUUGCAUGCACAUAUGUACAUAUUUGCACAAAUUGACUCUGCCCGGCCAC .(((((....((((((..((.((....)).)).........(((((..((((((((((((----((.((((......)))).))))))))))))))...)))))..))))))..))))). ( -40.00) >consensus AUGGCCAGCAGCAGUGAGGAGCCGAAGGUCUCAGCUAAUUAAAUUUCCCAGAUGUGUGUG____UGUGUGUUUGCAUACACAUAUUU____________AAAUUGGCUCUGCCAGGCCAC .(((((.((((.(((.(((((.(....).)))...................(((((((((..............)))))))))...................)).)))))))..))))). (-26.60 = -27.66 + 1.06)

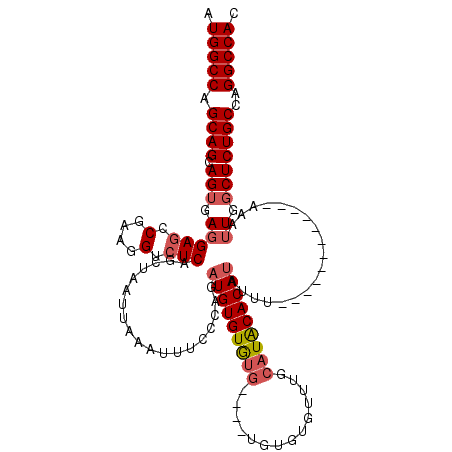
| Location | 2,351,499 – 2,351,605 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2351499 106 - 20766785 GUGGCCUGGCAGAGCCAAUUU------------AAAUAUGUGCAUGCAAACACACAC--ACACACACAUCUGGGAAAUUUAAUUAGUUGAGACCUUCGGCUCCUCACUGCUGCUGGCCAU ((((((.(((((..(((....------------.....((((..((......))..)--)))........)))..........((((.(((..(....)...)))))))))))))))))) ( -26.73) >DroSec_CAF1 1105 108 - 1 GUGGCCUGGCAGAGCCAAUUU------------AAAUAUGUGUAUGCAAACACACACACACACACACAUCUGGGAAAUUUAAUUAGCUGAGACCUUCGGCUCCUCACUGCUGCUGGCCAU ((((((.((((((((((....------------.....(((((.((......)).)))))..........)))...........(((((((...))))))).....)).))))))))))) ( -29.41) >DroSim_CAF1 1120 102 - 1 GUGGCCUGGCAGAGCCAAUUU------------AAAUAUGUGUAUGCAAACACACA------CACACAGCUGGGAAAUUUAAUUAGCUGAGACCUUCGGCUCCUCACUGCUGCUGGCCAU ((((((.(((((((((.....------------.....(((((.((......)).)------))))(((((((.........)))))))........))))).....))))...)))))) ( -34.30) >DroYak_CAF1 1120 116 - 1 GUGGCCGGGCAGAGUCAAUUUGUGCAAAUAUGUACAUAUGUGCAUGCAAACACCCA----CAUACACAUCUGGUAAAUUUAAUUAGCUGAGACCUUCGUUUCGUCCCUGCUGCUGGCCAU ((((((((((((((..((((((((((....))))))((((((..((....))..))----))))................))))..))(((((....)))))....))))..)))))))) ( -33.40) >consensus GUGGCCUGGCAGAGCCAAUUU____________AAAUAUGUGCAUGCAAACACACA____CACACACAUCUGGGAAAUUUAAUUAGCUGAGACCUUCGGCUCCUCACUGCUGCUGGCCAU ((((((.(((((((.........................(((........)))...............................(((((((...))))))).....)).))))))))))) (-22.26 = -22.20 + -0.06)

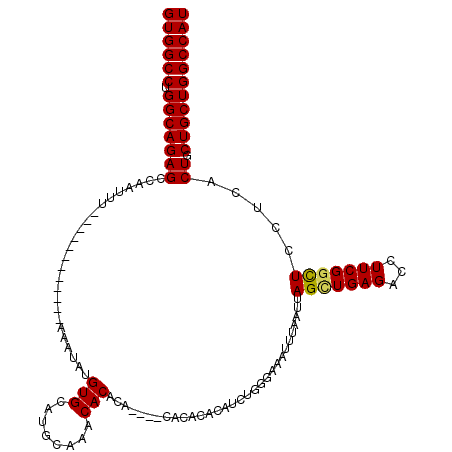
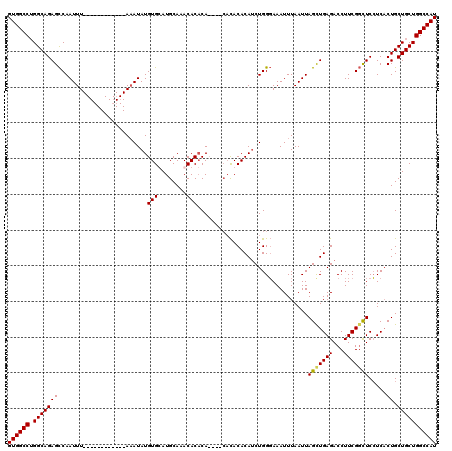
| Location | 2,351,539 – 2,351,634 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.958026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2351539 95 + 20766785 AAAUUUCCCAGAUGUGUGUGU--GUGUGUGUUUGCAUGCACAUAUUU------------AAAUUGGCUCUGCCAGGCCACAUAGCUGGCUAUGUAUGACUAUAUGACUA .......((((.(((((((((--(((((((....))))))))))...------------...(((((...)))))..)))))).)))).((((((....)))))).... ( -28.80) >DroSec_CAF1 1145 97 + 1 AAAUUUCCCAGAUGUGUGUGUGUGUGUGUGUUUGCAUACACAUAUUU------------AAAUUGGCUCUGCCAGGCCACAUAGCUGGCUAUAUAUGACUAUACGACUA .......((((.(((((((((((((((((....)))))))))))...------------...(((((...)))))..)))))).))))..................... ( -29.90) >DroSim_CAF1 1160 91 + 1 AAAUUUCCCAGCUGUGUG------UGUGUGUUUGCAUACACAUAUUU------------AAAUUGGCUCUGCCAGGCCACAUAGCUGGCUAUGUAUGACUAUACGACUA .......(((((((((((------((((((....)))))))......------------.....((((......))))))))))))))...(((((....))))).... ( -32.10) >DroYak_CAF1 1160 103 + 1 AAAUUUACCAGAUGUGUAUG----UGGGUGUUUGCAUGCACAUAUGUACAUAUUUGCACAAAUUGACUCUGCCCGGCCACAUCGCUGGCUGU--AUGACUAUAUGACUG .(((((..((((((((((((----((.((((......)))).))))))))))))))...))))).........((((((......)))))).--............... ( -27.00) >consensus AAAUUUCCCAGAUGUGUGUG____UGUGUGUUUGCAUACACAUAUUU____________AAAUUGGCUCUGCCAGGCCACAUAGCUGGCUAUGUAUGACUAUACGACUA .......((((.(((((.......(((((((......)))))))....................((((......))))))))).))))..................... (-18.48 = -18.98 + 0.50)

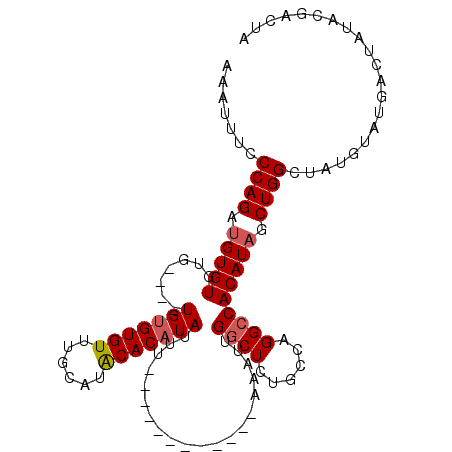
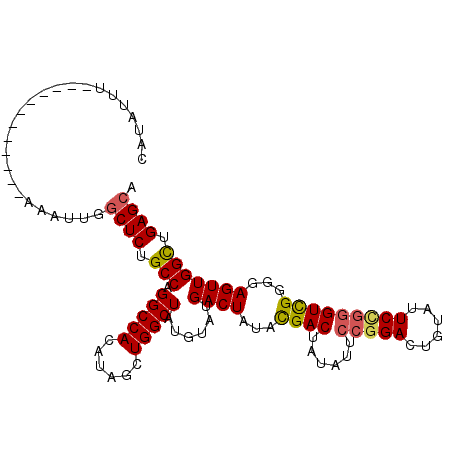
| Location | 2,351,577 – 2,351,674 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -26.41 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2351577 97 + 20766785 CAUAUUU------------AAAUUGGCUCUGCCAGGCCACAUAGCUGGCUAUGUAUGACUAUAUGACUAUAUUCCGGACUGCAUUCUGGGUCGGAGAGUUGGUUGAGCA .......------------.(((..(((((.((.((((((((((....))))))....................((((......)))))))))))))))..)))..... ( -28.10) >DroSec_CAF1 1185 97 + 1 CAUAUUU------------AAAUUGGCUCUGCCAGGCCACAUAGCUGGCUAUAUAUGACUAUACGACUAUAUUCCGGACUGUAUUCCGGGUCGGGGAGUUGGCUGAGCA .......------------......((((.(((.(((((......)))))......((((...((((......(((((......)))))))))...))))))).)))). ( -33.50) >DroSim_CAF1 1194 97 + 1 CAUAUUU------------AAAUUGGCUCUGCCAGGCCACAUAGCUGGCUAUGUAUGACUAUACGACUAUAUUCCGGACUGUAUUCCGGGUCGGGGAGUUGGCUGAGCA .......------------......((((.(((((.((((((((....)))))).........((((......(((((......))))))))).))..))))).)))). ( -34.10) >DroYak_CAF1 1196 103 + 1 CAUAUGUACAUAUUUGCACAAAUUGACUCUGCCCGGCCACAUCGCUGGCUGU--AUGACUAUAUGACUGUAU----GACUGUAUUCCGGGUUGGGGAGUUGGUUGAGCA ..............(((...(((..((((((((((((((......)))).((--(((..(((((....))))----)..)))))..)))))...)))))..)))..))) ( -27.40) >consensus CAUAUUU____________AAAUUGGCUCUGCCAGGCCACAUAGCUGGCUAUGUAUGACUAUACGACUAUAUUCCGGACUGUAUUCCGGGUCGGGGAGUUGGCUGAGCA .........................((((.(((.(((((......)))))......((((...((((......(((((......)))))))))...))))))).)))). (-26.41 = -26.60 + 0.19)

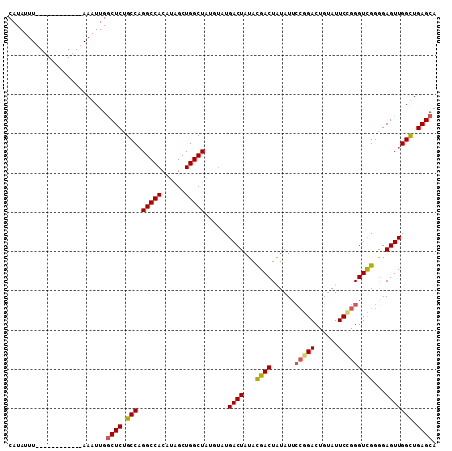
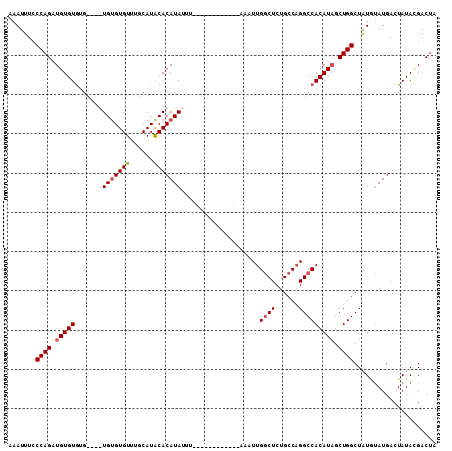
| Location | 2,351,674 – 2,351,794 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -35.08 |
| Energy contribution | -35.48 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2351674 120 + 20766785 AUGCACAUUUUUAAUUACGCUGCACAUUUGCAUUUGAAUUGCGCAUUUUACAGGCUGAUAAGCCGAAAUAAAGGCAACAGUUGUGCAGUGGCUGCCAAAGAUAUGUUGGCUCUGGCCACG .((((((...........((((.......(((.......)))((.(((((..((((....))))....)))))))..))))))))))(((((((((((.......)))))...)))))). ( -36.70) >DroSec_CAF1 1282 120 + 1 AUGCACAUUUUUAAUUAAGCUGCACAUUUGCAUUUGAAUUGCGCAUUUUACAGGCUGAUAAGCCGAAAUAAAGGCAACAGUUGUGCAGUGGCUGCCAAAGAUAUGUGGGCUCUGGCCACG .((((((.......(((((.((((....))))))))).....((.(((((..((((....))))....)))))))......))))))(((((((((...........)))...)))))). ( -34.60) >DroSim_CAF1 1291 120 + 1 AUGCACAUUUUUAAUUACGCUGCACAUUUGCAUUUGAAUUGCGCAUUUUACAGGCUGAUAAGCCGAAAUAAAGGCAACAGUUGUGCAGUGGCUGCCAAAGAUAUGUGGGCUCUGGCCACG ...(((((((((.....(((((((((...(((.......)))((.(((((..((((....))))....)))))))......))))))))).......)))).)))))(((....)))... ( -35.60) >DroEre_CAF1 1195 119 + 1 AUGCACAUUUUUAAUUACGCUGCACAUUUGCAUUUGAAUUGCGCAUUUUACAGGCUGAUAAGCCGAAAUAAAGGCAACAGUUGUGCAGUGGCUGCCAAAGAUAUGUUGGC-CUGGCCACG .((((((...........((((.......(((.......)))((.(((((..((((....))))....)))))))..))))))))))(((((((((((.......)))))-..)))))). ( -37.10) >DroYak_CAF1 1299 120 + 1 GUGCACAUUUUUAAUUACGCUGCACAUUUGCAUUUGAAUUGCGCAUUUUACAGGCUGAUAAGCCGAAAUAAAGGCAACAGUUGUGCAGUGGCUGCCAAAGAUAUGUUGGCUCUGGCCACG .((((((...........((((.......(((.......)))((.(((((..((((....))))....)))))))..))))))))))(((((((((((.......)))))...)))))). ( -36.60) >consensus AUGCACAUUUUUAAUUACGCUGCACAUUUGCAUUUGAAUUGCGCAUUUUACAGGCUGAUAAGCCGAAAUAAAGGCAACAGUUGUGCAGUGGCUGCCAAAGAUAUGUUGGCUCUGGCCACG .((((((...........((((.......(((.......)))((.(((((..((((....))))....)))))))..))))))))))(((((((((((.......)))))...)))))). (-35.08 = -35.48 + 0.40)

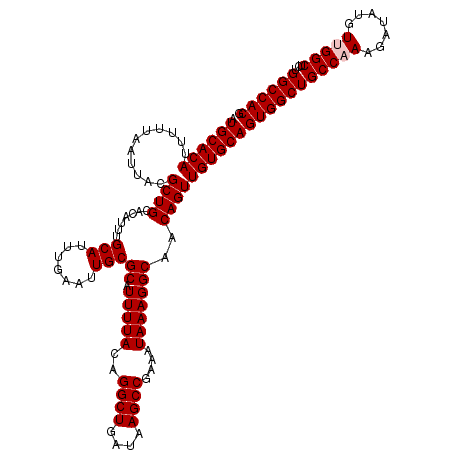
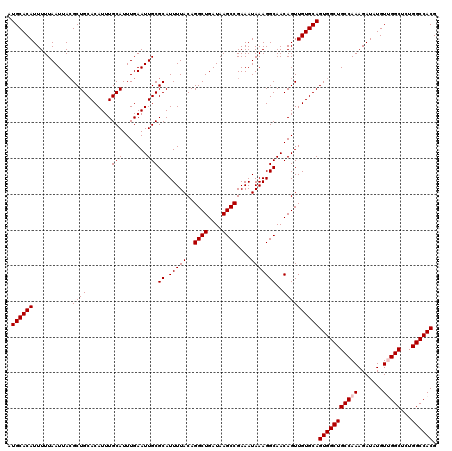
| Location | 2,351,674 – 2,351,794 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -31.96 |
| Energy contribution | -32.56 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2351674 120 - 20766785 CGUGGCCAGAGCCAACAUAUCUUUGGCAGCCACUGCACAACUGUUGCCUUUAUUUCGGCUUAUCAGCCUGUAAAAUGCGCAAUUCAAAUGCAAAUGUGCAGCGUAAUUAAAAAUGUGCAU .(((((....(((((.......))))).)))))((((((...(((((.(((((...((((....)))).))))).((((((.(((....).)).))))))..)))))......)))))). ( -34.60) >DroSec_CAF1 1282 120 - 1 CGUGGCCAGAGCCCACAUAUCUUUGGCAGCCACUGCACAACUGUUGCCUUUAUUUCGGCUUAUCAGCCUGUAAAAUGCGCAAUUCAAAUGCAAAUGUGCAGCUUAAUUAAAAAUGUGCAU .(((((....(((...........))).)))))((((((.(((..(((........)))....)))....(((..((((((.(((....).)).))))))..)))........)))))). ( -31.00) >DroSim_CAF1 1291 120 - 1 CGUGGCCAGAGCCCACAUAUCUUUGGCAGCCACUGCACAACUGUUGCCUUUAUUUCGGCUUAUCAGCCUGUAAAAUGCGCAAUUCAAAUGCAAAUGUGCAGCGUAAUUAAAAAUGUGCAU .(((((....(((...........))).)))))((((((...(((((.(((((...((((....)))).))))).((((((.(((....).)).))))))..)))))......)))))). ( -31.50) >DroEre_CAF1 1195 119 - 1 CGUGGCCAG-GCCAACAUAUCUUUGGCAGCCACUGCACAACUGUUGCCUUUAUUUCGGCUUAUCAGCCUGUAAAAUGCGCAAUUCAAAUGCAAAUGUGCAGCGUAAUUAAAAAUGUGCAU .(((((...-(((((.......))))).)))))((((((...(((((.(((((...((((....)))).))))).((((((.(((....).)).))))))..)))))......)))))). ( -36.30) >DroYak_CAF1 1299 120 - 1 CGUGGCCAGAGCCAACAUAUCUUUGGCAGCCACUGCACAACUGUUGCCUUUAUUUCGGCUUAUCAGCCUGUAAAAUGCGCAAUUCAAAUGCAAAUGUGCAGCGUAAUUAAAAAUGUGCAC .(((((....(((((.......))))).)))))((((((...(((((.(((((...((((....)))).))))).((((((.(((....).)).))))))..)))))......)))))). ( -34.50) >consensus CGUGGCCAGAGCCAACAUAUCUUUGGCAGCCACUGCACAACUGUUGCCUUUAUUUCGGCUUAUCAGCCUGUAAAAUGCGCAAUUCAAAUGCAAAUGUGCAGCGUAAUUAAAAAUGUGCAU .(((((....(((((.......))))).)))))((((((...(((((.(((((...((((....)))).))))).((((((.(((....).)).))))))..)))))......)))))). (-31.96 = -32.56 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:35 2006