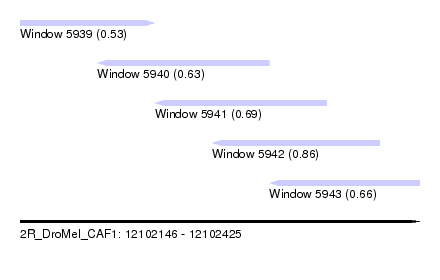
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,102,146 – 12,102,425 |
| Length | 279 |
| Max. P | 0.856369 |

| Location | 12,102,146 – 12,102,240 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -20.65 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12102146 94 + 20766785 UGCA--UUCUAUUUAGGCCAGGCCCACAAUGGUAUAGAAUAUCAGGGGUAGUACCAGGUUGUCCACCUGGUCGGUGAACGCCUCCACCAGAGCCGA ....--.........((((..((((....((((((...)))))).)))).(.(((((((.....))))))))((((........)))).).))).. ( -29.80) >DroPse_CAF1 12031 96 + 1 AGGAGAUCAGAAUCAGGCCAGGCCAACGAGUAUAUAAAAGACUAGGGGCAGCACCAGAUUGUCCACCUGGUCGGUGAAGGCCUCCACCAGGGCUGA ............((((.((....................(((((((((((((....).))).)).)))))))((((........)))).)).)))) ( -29.30) >DroEre_CAF1 6752 93 + 1 UGCA--UCC-AAUUAGGCCAGGCCCACAAUGGUAUAGAAUAUCAGGGGCAGCACCAGGUUGUCCACCUGGUCGGUGAAUGCUUCCAACAGAGCCGA .(((--(.(-(....(((((((.......((((((...)))))).(((((((.....))))))).)))))))..)).))))............... ( -30.20) >DroYak_CAF1 6835 93 + 1 UGGA--UAC-AAUUAGGCCAGGCCCACAAUGGUAUAGAAUAUCAGGGGCAGGACCAGGUUGUCCACCUGGUCGGUGAACGCCUCCACCAAAGCAGA ((((--...-.....((((((((((....((((((...)))))).)))).((((......))))..))))))(((....))))))).......... ( -33.00) >DroAna_CAF1 2623 93 + 1 AUCA--ACG-CUUCAGGCUAGGCCAACGAUGAUGUAGAAUAUCAGGGGCAGCACCAGGUUGUCCACCUGGUCGGUGAAAGCCUCUACCAAAGCCGA ....--...-.....((((.(((((....((((((...)))))).(((((((.....)))))))...)))))(((....)))........)))).. ( -30.60) >DroPer_CAF1 11777 96 + 1 AGGAGAUCAGAAUCAGGCCAGGCCAACGAGUAUAUAAAAGACUAGGGGCAGCACCAGAUUGUCCACCUGGUCGGUGAAGGCCUCCACCAGGGCUGA ............((((.((....................(((((((((((((....).))).)).)))))))((((........)))).)).)))) ( -29.30) >consensus AGCA__UCC_AAUCAGGCCAGGCCAACAAUGAUAUAGAAUAUCAGGGGCAGCACCAGGUUGUCCACCUGGUCGGUGAACGCCUCCACCAGAGCCGA ...............(((((((.......(((((.....))))).((((((.......)))))).)))))))((((........))))........ (-20.65 = -21.57 + 0.92)

| Location | 12,102,200 – 12,102,320 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -49.80 |
| Consensus MFE | -29.11 |
| Energy contribution | -28.59 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12102200 120 - 20766785 CCGUCUGGCUCCUGGAGAUUUCCGGCCUGGUGGCCAUGAGCCAGGCCAAGUGGUUCGCCACCAUUUUCGCAGCACUGAAUUCGGCUCUGGUGGAGGCGUUCACCGACCAGGUGGACAACC ..(((((((((..((......))((((....))))..))))))))).....((((.(((.((((.....(((..(((....)))..))))))).)))(((((((.....))))))))))) ( -53.50) >DroVir_CAF1 3184 120 - 1 CCGUUUGGCUGCUGCAGCUGGGCGGAGUUCUGGUCAUGAGCCAGGCCAAAUGGUUCGCCACGCUGUUUGCGGCGCUCAACACGGCUUUGGUGGAGGCGUCUACCGAUCAGGUGGACAAUC (((((..((((...))))..)))))....(((((.....)))))(((......(((((((.((((....))))(((......)))..))))))))))(((((((.....))))))).... ( -51.40) >DroPse_CAF1 12087 120 - 1 CCGUCUGGCUGCUGGAGGCGACCGGUCUGCUGGCCAUGACACAGGCCAAGUGGUUCGCAACCAUUUUCGCAGCCAUCAACUCAGCCCUGGUGGAGGCCUUCACCGACCAGGUGGACAAUC ((..((((.((.(((((((....((.((((.((((........))))(((((((.....)))))))..)))))).....(((.((....)).)))))))))).)).))))..))...... ( -51.40) >DroMoj_CAF1 3250 120 - 1 CCGUCGGGCUGCUGCAGCUGAGCGGCGUCCUUGUCAUGACCCAGGCGAAAUGGUUCGCCACGAUUUUUGCCACACUUAAUGCAGCUCUAGUUGAGGCACUUACCGACCAGGUGGACAAUC ..(((((..((((.(((((((((((.(((........))))).((((((((((....)))....)))))))............)))).))))).))))....)))))............. ( -41.20) >DroAna_CAF1 2676 120 - 1 CCAUUUGGCUACUGGACGCGUUCGGUCUGGUGGCGAUGACCCAGGCCAAGUGGUUCGCCUCGGUCUUUGCCGCUUUGAACUCGGCUUUGGUAGAGGCUUUCACCGACCAGGUGGACAACC ((((((((((((..((((.((((((...((((((((.((((.((((..........)))).)))).)))))))))))))).))..))..)))).((......))..))))))))...... ( -49.90) >DroPer_CAF1 11833 120 - 1 CCGUCUGGCUGCUGGAGGCGACCGGUCUGCUGGCCAUGACACAGGCCAAGUGGUUCGCAACCAUUUUCGCAGCCAUCAACUCAGCCCUGGUGGAGGCCUUCACCGACCAGGUGGACAAUC ((..((((.((.(((((((....((.((((.((((........))))(((((((.....)))))))..)))))).....(((.((....)).)))))))))).)).))))..))...... ( -51.40) >consensus CCGUCUGGCUGCUGGAGGCGACCGGUCUGCUGGCCAUGACCCAGGCCAAGUGGUUCGCCACCAUUUUCGCAGCACUCAACUCAGCUCUGGUGGAGGCCUUCACCGACCAGGUGGACAAUC ((((((((..(((((......)))))....(((((........)))))..((((..(((.(((((...((.............))...))))).)))....)))).))))))))...... (-29.11 = -28.59 + -0.52)


| Location | 12,102,240 – 12,102,360 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -48.24 |
| Consensus MFE | -25.72 |
| Energy contribution | -24.58 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12102240 120 - 20766785 CUGGAGGGCACCAUUGCGUUCGUGGUGUCCAUUCUCAUGGCCGUCUGGCUCCUGGAGAUUUCCGGCCUGGUGGCCAUGAGCCAGGCCAAGUGGUUCGCCACCAUUUUCGCAGCACUGAAU ((((.(((((((((.......)))))))))...(((((((((..(.((((...((......)))))).)..)))))))))))))((.(((((((.....)))))))..)).......... ( -54.80) >DroVir_CAF1 3224 120 - 1 CUGGAGGGCACCGUGGCCUUUGUGCUGUCCGUGCUGCUCUCCGUUUGGCUGCUGCAGCUGGGCGGAGUUCUGGUCAUGAGCCAGGCCAAAUGGUUCGCCACGCUGUUUGCGGCGCUCAAC ..(((.(((((.(......).))))).)))(((((((.(((((((..((((...))))..)))))))...((((...((((((.......))))))))))........)))))))..... ( -54.20) >DroPse_CAF1 12127 120 - 1 CUGGAAGGCACUAUCGCCUUCGUGAUGUCCAUCCUUCUGUCCGUCUGGCUGCUGGAGGCGACCGGUCUGCUGGCCAUGACACAGGCCAAGUGGUUCGCAACCAUUUUCGCAGCCAUCAAC ...((((((......))))))..((((..((......))..))))((((((((((..(((((((.(....(((((........)))))).))).))))..))).....)))))))..... ( -46.90) >DroMoj_CAF1 3290 120 - 1 CUGGAGGGCACUGUUGCCUUUGUGCUGUCCAUCCUGCUGUCCGUCGGGCUGCUGCAGCUGAGCGGCGUCCUUGUCAUGACCCAGGCGAAAUGGUUCGCCACGAUUUUUGCCACACUUAAU .((((.(((((..........))))).))))....((.....(((((((.(((((......))))))))).............((((((....))))))..)))....)).......... ( -39.00) >DroAna_CAF1 2716 120 - 1 UUGGAGGGCACCAUUGCCUUUGUGGCUUCCAUCUUGCUCUCCAUUUGGCUACUGGACGCGUUCGGUCUGGUGGCGAUGACCCAGGCCAAGUGGUUCGCCUCGGUCUUUGCCGCUUUGAAC .(((((((((.....(((.....)))........)))))))))...(((.((..(((.(....))))..))(((((.((((.((((..........)))).)))).))))))))...... ( -47.62) >DroPer_CAF1 11873 120 - 1 CUGGAAGGCACUAUCGCCUUCGUGAUGUCCAUCCUUCUGUCCGUCUGGCUGCUGGAGGCGACCGGUCUGCUGGCCAUGACACAGGCCAAGUGGUUCGCAACCAUUUUCGCAGCCAUCAAC ...((((((......))))))..((((..((......))..))))((((((((((..(((((((.(....(((((........)))))).))).))))..))).....)))))))..... ( -46.90) >consensus CUGGAGGGCACCAUUGCCUUCGUGAUGUCCAUCCUGCUGUCCGUCUGGCUGCUGGAGGCGACCGGUCUGCUGGCCAUGACCCAGGCCAAGUGGUUCGCCACCAUUUUCGCAGCACUCAAC ...((((((......))))))((((...((((..................(((((......)))))....(((((........))))).)))).))))...................... (-25.72 = -24.58 + -1.13)
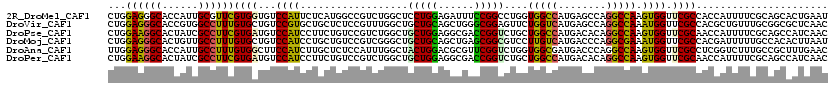
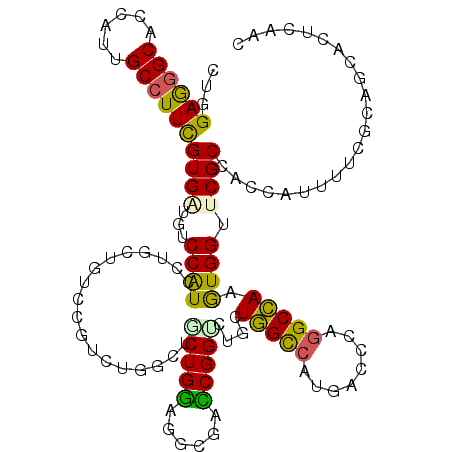
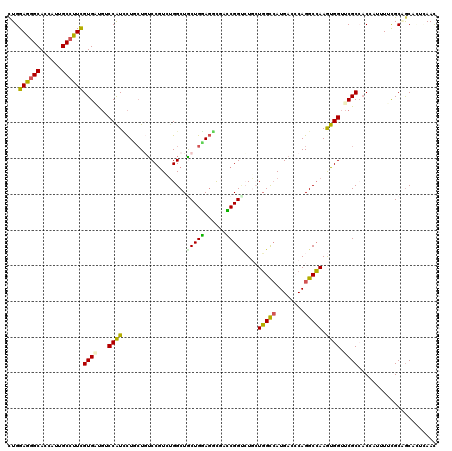
| Location | 12,102,280 – 12,102,397 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.45 |
| Mean single sequence MFE | -45.43 |
| Consensus MFE | -33.29 |
| Energy contribution | -35.35 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12102280 117 - 20766785 UAACACGCGUGCCAUACUUCUAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCGUUCGUGGUGUCCAUUCUCAUGGCCGUCUGGCUCCUGGAGAUUUCCGGCCUGGUGGCCAUGAG ..................(((((((((.....))).))))))(((((((((.......)))))))))...(((((((((..(.((((...((......)))))).)..))))))))) ( -50.80) >DroSec_CAF1 6514 117 - 1 UAAUACAUGUGUCAUACUUCUAGGAUCUUCCCGCUCCCUGGAGGGCACCAUUGCGUUCGUGGUGUCCAUUCUCAUGGCCGUCUGGCUGCUGGCGAUUUCCGGCCUGGUGGCCAUGAG ..................((((((............))))))(((((((((.......)))))))))...((((((((((.(..(..(((((......))))))..))))))))))) ( -47.40) >DroSim_CAF1 6479 117 - 1 UAACACAUGUGUCAUACUUCUAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCGUUCGUGGUGUCCAUUCUCAUGGCCGCCUGGCUGCUGGAGAUUUCCGGCCUGGUGGCCAUGAG ..(((....)))......(((((((((.....))).))))))(((((((((.......)))))))))...((((((((((((.((((...((......)))))).)))))))))))) ( -55.70) >DroEre_CAF1 6885 117 - 1 UAAUACAUGUGUCAUACCCUUAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCUUUCGUCGUCUCCAUUCUAACAGCCGUCUGGCUGCUGGAGGCUACCGGCUUGGUGGCCAUGAG .....((((.(((...(((((.(((((.....)))))...))))).((((..(((...((.(((((((......(((((....))))).))))))).)).))).))))))))))).. ( -45.30) >DroYak_CAF1 6968 117 - 1 GAAUACAUGUGUCGUACUUUCAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCGUUCGUCGUGUCCAUUCUUAUAGCCGUCUGGCUGCUGGAGACCACCGGCUUGGUGGCCAUGAG (((..((.(((..((.(((((((((((.....))).))))))))))..)))))..))).(((((.(((((.....((((....))))(((((......)))))..))))).))))). ( -40.20) >DroPer_CAF1 11913 108 - 1 ---------UUCAACUCUUGCAGAUUCCUCACGUUCCCUGGAAGGCACUAUCGCCUUCGUGAUGUCCAUCCUUCUGUCCGUCUGGCUGCUGGAGGCGACCGGUCUGCUGGCCAUGAC ---------.((..((((.((((...((..(((..(...(((.((.((.(((((....))))))))).)))....)..)))..)))))).))))..))..((((....))))..... ( -33.20) >consensus UAAUACAUGUGUCAUACUUCUAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCGUUCGUGGUGUCCAUUCUCAUGGCCGUCUGGCUGCUGGAGACUACCGGCCUGGUGGCCAUGAG ..................(((((((((.....))).))))))(((((((((.......)))))))))...((((((((((((.((((...((......)))))).)))))))))))) (-33.29 = -35.35 + 2.06)

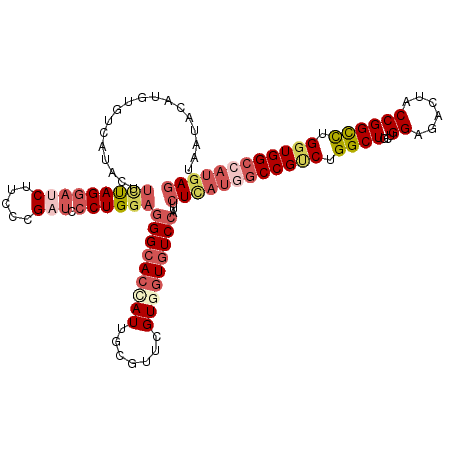
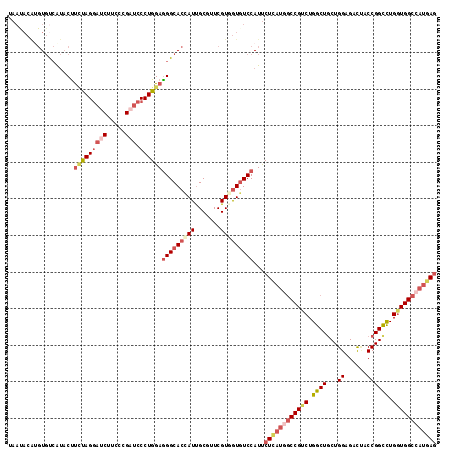
| Location | 12,102,320 – 12,102,425 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.88 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
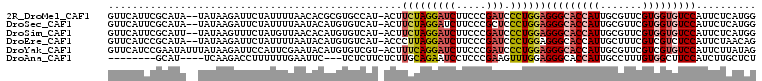
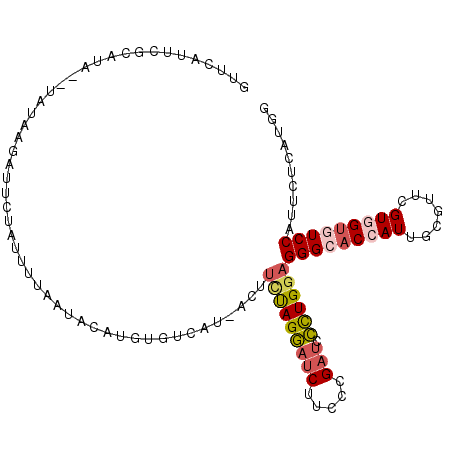
>2R_DroMel_CAF1 12102320 105 - 20766785 GUUCAUUCGCAUA--UAUAAGAUUCUAUUUUAACACGCGUGCCAU-ACUUCUAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCGUUCGUGGUGUCCAUUCUCAUGG .......(((...--..((((((...))))))....)))..((((-...(((((((((.....))).))))))(((((((((.......)))))))))......)))) ( -28.30) >DroSec_CAF1 6554 105 - 1 GUUCAUUCGCAUA--UAUAAGAUUCUAUUUUAAUACAUGUGUCAU-ACUUCUAGGAUCUUCCCGCUCCCUGGAGGGCACCAUUGCGUUCGUGGUGUCCAUUCUCAUGG ((.....(((((.--(((.(((......))).))).)))))....-)).((((((............))))))(((((((((.......))))))))).......... ( -25.60) >DroSim_CAF1 6519 105 - 1 GUUCAUUCGCAUU--UAUAAGUUUCUAUGUUAACACAUGUGUCAU-ACUUCUAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCGUUCGUGGUGUCCAUUCUCAUGG .............--.....(((........))).((((.(....-...(((((((((.....))).))))))(((((((((.......)))))))))...).)))). ( -27.50) >DroEre_CAF1 6925 105 - 1 GUUCAUCCGCAUA--UAUAAGAUUCUAUUUUAAUACAUGUGUCAU-ACCCUUAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCUUUCGUCGUCUCCAUUCUAACAG .......(((((.--(((.(((......))).))).)))))....-.(((((.(((((.....)))))...)))))................................ ( -18.20) >DroYak_CAF1 7008 107 - 1 GUUCAUCCGAAUAUUUAUAAGAUUCCAUUCGAAUACAUGUGUCGU-ACUUUCAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCGUUCGUCGUGUCCAUUCUUAUAG ((((....))))...(((((((...(((.((((..((.(((..((-.(((((((((((.....))).))))))))))..)))))..))))..))).....))))))). ( -27.30) >DroAna_CAF1 2796 93 - 1 --------GCAU----UCAAGACCUUUUUUGAAUUC---UCUCUUCUCUUGCAGAAUCCUCCCGAAGUUUGGAGGGCACCAUUGCCUUUGUGGCUUCCAUCUUGCUCU --------((((----((((((....)))))))...---........................((((((..(((((((....)))))))..)))))).....)))... ( -20.20) >consensus GUUCAUUCGCAUA__UAUAAGAUUCUAUUUUAAUACAUGUGUCAU_ACUUCUAGGAUCUUCCCGAUCCCUGGAGGGCACCAUUGCGUUCGUGGUGUCCAUUCUCAUGG .................................................(((((((((.....))).))))))(((((((((.......))))))))).......... (-16.19 = -16.88 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:53 2006