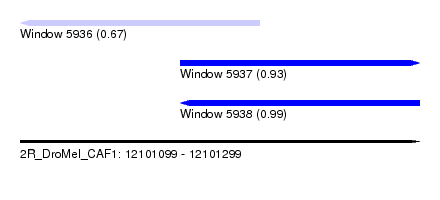
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,101,099 – 12,101,299 |
| Length | 200 |
| Max. P | 0.994085 |

| Location | 12,101,099 – 12,101,219 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -50.38 |
| Consensus MFE | -41.58 |
| Energy contribution | -42.38 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12101099 120 - 20766785 UCCUUGAUGACCUUGGUCAGACGCUCGGCGUCGACCUCAACGCCCACAGAGCUGAGGAUCUUCUCCAGAUCGCUGUUGGCGGGCGAGUCCUUGCCACCGAGGACGGCCAGAAGGUAAGCA ....((...(((((((((.(((((...)))))))))....((((.((((..(((..(.....)..)))....)))).))))(((..(((((((....))))))).)))..)))))...)) ( -50.90) >DroSec_CAF1 5393 120 - 1 UCCUUGAUGACCUUGGUCAGACGCUCGGCGUCAACCUCAACGCCCACGGAGCUGAGGAUCUUCUCCAGAUCGCCGUUGGCGGGCGAGUCCUUGCCACCGAGGACGGCCAGAAGGUAAGCA ....((...(((((((((.(((((...)))))..((((..((((.((((..(((..(.....)..)))....)))).))))(((((....)))))...))))..)))))..))))...)) ( -51.20) >DroSim_CAF1 5374 120 - 1 UCCUUGAUGACCUUGGUCAGACGCUCGGCGUCGACCUCAACGCCCACGGAGCUGAGGAUCUUCUCCAGAUCGCCGUUGGCGGGCGAGUCCUUGCCACCGAGGACGGCCAGAACGUAAGCA ..(((.(((.....((((.(((((...)))))))))((..((((.((((..(((..(.....)..)))....)))).))))(((..(((((((....))))))).))).)).)))))).. ( -48.10) >DroEre_CAF1 5610 120 - 1 UCCUUGAUGACCUUGGUCAGGCGCUCGGUGUCGACCUCGAUGCCCACAGAGCUGAGGAUCUUCUCCAGGUCGGCGUUGGCGGGGGAGUCCUUGCCACCGAGGACGGCCAGAAGGUAAGCA .((((..((((....))))...(((((((((((....)))))))....)))).))))(((((((...(((((.((.((((((((....)))))))).))....))))))))))))..... ( -51.20) >DroYak_CAF1 5782 120 - 1 UCCUUGAUGACCUUGGUCAGGCGCUCAGCGUCGACCUCAAUGCCCACAGAGCUGAGGAUCUUCUCCAGGUCGGCGUUGGCGGGGGUGUCCUUGCCACCGAGGACGGCCAGAAGGUAAGCA .((((..((((....))))..(((.((((((((((((....((.......))...(((.....)))))))))))))))))).((.((((((((....)))))))).))..))))...... ( -50.50) >consensus UCCUUGAUGACCUUGGUCAGACGCUCGGCGUCGACCUCAACGCCCACAGAGCUGAGGAUCUUCUCCAGAUCGCCGUUGGCGGGCGAGUCCUUGCCACCGAGGACGGCCAGAAGGUAAGCA ....((...(((((((((.(((((...)))))..((((..((((.((((..(((..(.....)..)))....)))).))))(((((....)))))...))))..)))))..))))...)) (-41.58 = -42.38 + 0.80)

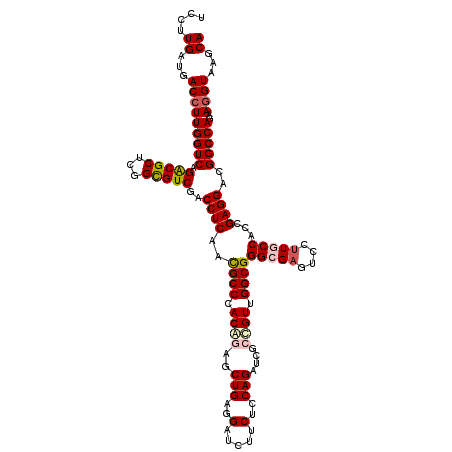
| Location | 12,101,179 – 12,101,299 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -52.50 |
| Consensus MFE | -45.92 |
| Energy contribution | -46.36 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12101179 120 + 20766785 UUGAGGUCGACGCCGAGCGUCUGACCAAGGUCAUCAAGGAGCUGGCUGGCAAGAGCAUCGACGACCUGAUCAAGGAGGGUCGCGAGAAGCUCUCCUCGAUGCCGGUGGGCGGCGGUGGUG ....(((((((((...))))).)))).....(((((..(.(((.((((((((((((.(((.((((((.........)))))))))...)))))......))))))).)))..)..))))) ( -53.60) >DroSec_CAF1 5473 120 + 1 UUGAGGUUGACGCCGAGCGUCUGACCAAGGUCAUCAAGGAGCUGGCUGGCAAGAGCAUCGACGACCUGAUCAAGGAGGGUCGCGAGAAGCUCUCCUCGAUGCCGGUGGGCGGCGGUGGUG ....(((((((((...))))).)))).....(((((..(.(((.((((((((((((.(((.((((((.........)))))))))...)))))......))))))).)))..)..))))) ( -51.50) >DroSim_CAF1 5454 120 + 1 UUGAGGUCGACGCCGAGCGUCUGACCAAGGUCAUCAAGGAGCUGGCUGGCAAGAGCAUCGACGACCUGAUCAAGGAGGGUCGCGAGAAGCUCUCCUCGAUGCCGGUGGGCGGCGGUGGUG ....(((((((((...))))).)))).....(((((..(.(((.((((((((((((.(((.((((((.........)))))))))...)))))......))))))).)))..)..))))) ( -53.60) >DroEre_CAF1 5690 120 + 1 UCGAGGUCGACACCGAGCGCCUGACCAAGGUCAUCAAGGAGCUGGCUGGCAAGAGCAUCGAGGACCUCAUCAAGGAGGGACGCGAGAAGCUCUCCUCGAUGCCGGUGGGCGGCGGUGGCG (((....)))(((((..(((((.(((..((.((((.(((((((....)))..((((.(((.(..((((......))))..).)))...)))))))).)))))))))))))).)))))... ( -55.20) >DroYak_CAF1 5862 120 + 1 UUGAGGUCGACGCUGAGCGCCUGACCAAGGUCAUCAAGGAGCUGGCUGGCAAGAGCAUCGAGGAGCUCAUCAAGCAGGGACGCGAGAAGCUCUCCUCGAUGCCGGUGGGCGGCGGUGGUG ..........((((.(.((((((((....)))).......(((.(((((((......((((((((..(.((..((......))..)).)..))))))))))))))).))))))).))))) ( -48.60) >consensus UUGAGGUCGACGCCGAGCGUCUGACCAAGGUCAUCAAGGAGCUGGCUGGCAAGAGCAUCGACGACCUGAUCAAGGAGGGUCGCGAGAAGCUCUCCUCGAUGCCGGUGGGCGGCGGUGGUG ..........(((((..(((((.(((..((.((((.(((((((....)))..((((.(((.((((((.........)))))))))...)))))))).)))))))))))))).)))))... (-45.92 = -46.36 + 0.44)


| Location | 12,101,179 – 12,101,299 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -40.54 |
| Energy contribution | -41.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12101179 120 - 20766785 CACCACCGCCGCCCACCGGCAUCGAGGAGAGCUUCUCGCGACCCUCCUUGAUCAGGUCGUCGAUGCUCUUGCCAGCCAGCUCCUUGAUGACCUUGGUCAGACGCUCGGCGUCGACCUCAA .......((((.....))))(((((((((.((.....))....))))))))).((((((((((.(((..........)))...)))))))))).((((.(((((...))))))))).... ( -45.80) >DroSec_CAF1 5473 120 - 1 CACCACCGCCGCCCACCGGCAUCGAGGAGAGCUUCUCGCGACCCUCCUUGAUCAGGUCGUCGAUGCUCUUGCCAGCCAGCUCCUUGAUGACCUUGGUCAGACGCUCGGCGUCAACCUCAA .((((..((((.....))))(((((((((.((.....))....))))))))).((((((((((.(((..........)))...))))))))))))))..(((((...)))))........ ( -42.70) >DroSim_CAF1 5454 120 - 1 CACCACCGCCGCCCACCGGCAUCGAGGAGAGCUUCUCGCGACCCUCCUUGAUCAGGUCGUCGAUGCUCUUGCCAGCCAGCUCCUUGAUGACCUUGGUCAGACGCUCGGCGUCGACCUCAA .......((((.....))))(((((((((.((.....))....))))))))).((((((((((.(((..........)))...)))))))))).((((.(((((...))))))))).... ( -45.80) >DroEre_CAF1 5690 120 - 1 CGCCACCGCCGCCCACCGGCAUCGAGGAGAGCUUCUCGCGUCCCUCCUUGAUGAGGUCCUCGAUGCUCUUGCCAGCCAGCUCCUUGAUGACCUUGGUCAGGCGCUCGGUGUCGACCUCGA ((.(((((.((((.((((((((((((((((((...(((.(..((((......))))..).))).))))..((......))))))))))).))..)))..))))..))))).))....... ( -50.30) >DroYak_CAF1 5862 120 - 1 CACCACCGCCGCCCACCGGCAUCGAGGAGAGCUUCUCGCGUCCCUGCUUGAUGAGCUCCUCGAUGCUCUUGCCAGCCAGCUCCUUGAUGACCUUGGUCAGGCGCUCAGCGUCGACCUCAA .......((((.....))))...((((.((((((.(((.((....)).))).)))))).((((((((..((((((....))......((((....))))))))...)))))))))))).. ( -43.90) >consensus CACCACCGCCGCCCACCGGCAUCGAGGAGAGCUUCUCGCGACCCUCCUUGAUCAGGUCGUCGAUGCUCUUGCCAGCCAGCUCCUUGAUGACCUUGGUCAGACGCUCGGCGUCGACCUCAA .......((((.....))))(((((((((.((.....))....))))))))).((((((((((.(((..........)))...)))))))))).((((.(((((...))))))))).... (-40.54 = -41.14 + 0.60)

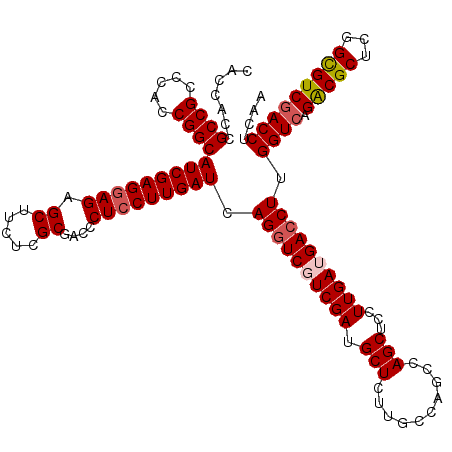
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:48 2006