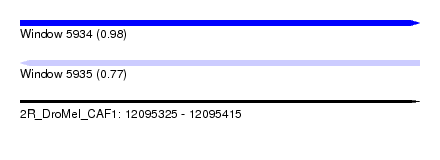
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,095,325 – 12,095,415 |
| Length | 90 |
| Max. P | 0.980968 |

| Location | 12,095,325 – 12,095,415 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -13.38 |
| Energy contribution | -16.62 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.980968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
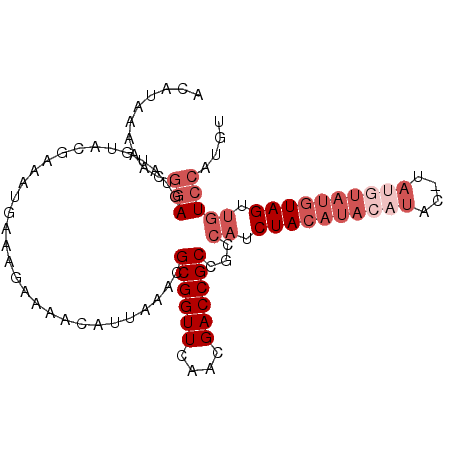
>2R_DroMel_CAF1 12095325 90 + 20766785 ACAUAAAAACGGAGUAUGUACGA---------AAACAUUAAACGCGGUUCAACGACCGCCGCCAUCUACAUACAUACAUACAUAUGUAGUUGUCCAUGU ..........((((((((((.(.---------...).......((((((....)))))).......))))))).(((((....)))))....))).... ( -19.10) >DroSec_CAF1 8593 97 + 1 ACAUAAAAACGGAGUAUGUACGAAAUGAAAGAAAAUAUUAAACGCGGUUCAACGACCGCCGCCAUCUACAUACAUG--UAUGUAUGUAGUUGUCCAUGU ..........(((..............................((((((....))))))...((.(((((((((..--..))))))))).))))).... ( -22.90) >DroSim_CAF1 10126 96 + 1 ACAUAAAAACGGAGUAUGUACGAAAUGAAAGAAAAUAUUAAACGCGGUUCAACGACCGCCGCCAUCUACAUACAUACAUAUGUAUGUAG---UCCAUGU ..........(((..............................((((((....))))))......((((((((((....))))))))))---))).... ( -22.90) >DroEre_CAF1 8579 90 + 1 ACAUUAAACUACAGUGUGUAC-AAAUGAACAAAAACAUACAACGCGGUUCAACGACCGCCUCCAUCUACAUUCACAC--------AUAGUUGUCCAUGU ((((..(((((..(((((...-..(((........))).....((((((....)))))).............)))))--------.)))))....)))) ( -17.30) >consensus ACAUAAAAACGGAGUAUGUACGAAAUGAAAGAAAACAUUAAACGCGGUUCAACGACCGCCGCCAUCUACAUACAUAC_UAUGUAUGUAGUUGUCCAUGU ..........(((..............................((((((....))))))...((.((((((((((....)))))))))).))))).... (-13.38 = -16.62 + 3.25)

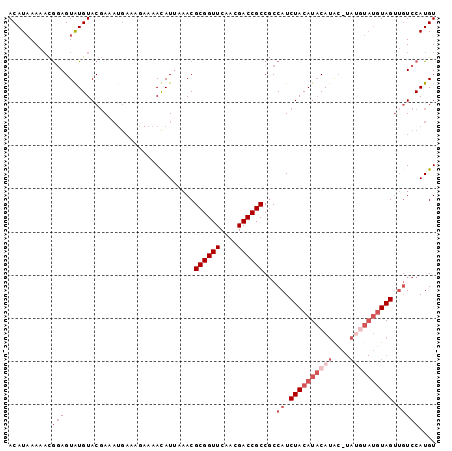
| Location | 12,095,325 – 12,095,415 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -17.59 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
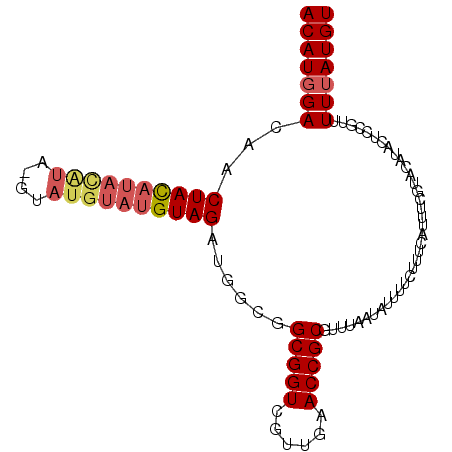
>2R_DroMel_CAF1 12095325 90 - 20766785 ACAUGGACAACUACAUAUGUAUGUAUGUAUGUAGAUGGCGGCGGUCGUUGAACCGCGUUUAAUGUUU---------UCGUACAUACUCCGUUUUUAUGU (((((((...((((((((((....))))))))))((((.((((((......))))).....((((..---------....)))).).)))).))))))) ( -24.80) >DroSec_CAF1 8593 97 - 1 ACAUGGACAACUACAUACAUA--CAUGUAUGUAGAUGGCGGCGGUCGUUGAACCGCGUUUAAUAUUUUCUUUCAUUUCGUACAUACUCCGUUUUUAUGU ..(((((((.(((((((((..--..))))))))).))...(((((......)))))..............................)))))........ ( -23.60) >DroSim_CAF1 10126 96 - 1 ACAUGGA---CUACAUACAUAUGUAUGUAUGUAGAUGGCGGCGGUCGUUGAACCGCGUUUAAUAUUUUCUUUCAUUUCGUACAUACUCCGUUUUUAUGU (((((((---((((((((((....))))))))))((((.((((((......)))))((......................))...).)))).))))))) ( -24.05) >DroEre_CAF1 8579 90 - 1 ACAUGGACAACUAU--------GUGUGAAUGUAGAUGGAGGCGGUCGUUGAACCGCGUUGUAUGUUUUUGUUCAUUU-GUACACACUGUAGUUUAAUGU ((((....((((((--------(((((..((((((((((.(((((......)))))....((......)))))))))-)))))))).))))))..)))) ( -21.70) >consensus ACAUGGACAACUACAUACAUA_GUAUGUAUGUAGAUGGCGGCGGUCGUUGAACCGCGUUUAAUAUUUUCUUUCAUUUCGUACAUACUCCGUUUUUAUGU (((((((...((((((((((....))))))))))......(((((......)))))....................................))))))) (-17.59 = -18.90 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:44 2006