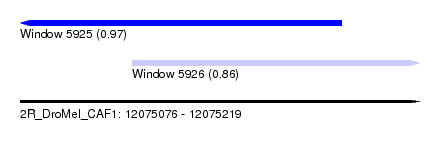
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,075,076 – 12,075,219 |
| Length | 143 |
| Max. P | 0.966833 |

| Location | 12,075,076 – 12,075,191 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.23 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -24.46 |
| Energy contribution | -23.43 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
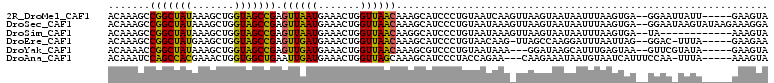
>2R_DroMel_CAF1 12075076 115 - 20766785 UACUUAAC----UUGAUUA-CAGGGAUGCUUUGUUAACCAGUUUCAUUAACUCGGCUACCAGCUUUAUAGCCGGCUUUGUGAUAUUCUCCGUGCUGGGUUACAUGGCCCACACACUGGGU .......(----(((....-)))).............(((((.((((.((..((((((.........))))))..)).))))........(((.((((((....)))))))))))))).. ( -31.70) >DroVir_CAF1 104776 116 - 1 ---UGAGCUAUGUAUUUUA-CAGAGAUGCUCUGCUUACCAGUUUUAUAAACUCGGCCACGAGUUUUGUGGCCGGCUUUGUGAUCUUUUCUGUGCUGGGUUACAUGGCGCACACAUCGGGC ---((.((((((((.....-(((((...)))))....(((((.(((((((..(((((((((...)))))))))..)))))))..........)))))..)))))))).)).......... ( -40.80) >DroGri_CAF1 91763 119 - 1 UGAUUAACUGUGUGUAUUA-CAGAGAUGCCCUGCUUACAAGUUUUAUAAACUCUGCAACGAGUUUUGUGGCGGGCUUUGUGAUAUUCUCCGUGCUGGGCUAUAUGGCACAUACUUCCGGC ......((.(.(.((((((-(((((....((((((.((((........(((((......)))))))))))))))))))))))))).).).))((((((...((((...))))..)))))) ( -31.70) >DroEre_CAF1 70790 114 - 1 UGGCUAA-----CUUGUUA-CAGGGAUGCUUUGUUAACCAGUUUCAUCAACUCGGCUACCAGCUUCAUAGCCGGCUUUGUGAUAUUCUCCGUGCUGGGUUACAUGGCCCACACCUUGGGU ((((...-----...))))-(((((..(((.(((...(((((.((((.((..((((((.........))))))..)).))))..........)))))...))).))).....)))))... ( -29.60) >DroWil_CAF1 102294 110 - 1 U----AU------CUAUUACUAGAGAUGCUUUGCUCACCAGCUUCAUUAAUUCGGCUACAAGUUUUGUGGCUGGUUUUGUGAUAUUCUCCGUAUUGGGUUAUAUGGCUCAUACAUUGGGU .----((------(((.(((..((((......(((....))).((((....(((((((((.....)))))))))....))))...)))).))).)))))......(((((.....))))) ( -27.40) >DroAna_CAF1 66374 112 - 1 UUCUUG-------UUCUGG-UAGGGAUGCUUUGCUAACCAGUUUCAUCAAUUCAGCCACCAGUUUCGUGGCUGGAUUUGUGAUAUUCUCGGUGCUGGGCUACAUGGCCCACACAUUGGGA ......-------..((((-(((.((....)).)).)))))..((((.(((((((((((.......))))))))))).))))...((((((((.((((((....))))))..)))))))) ( -44.50) >consensus U__UUAA_____UUUAUUA_CAGAGAUGCUUUGCUAACCAGUUUCAUAAACUCGGCUACCAGUUUUGUGGCCGGCUUUGUGAUAUUCUCCGUGCUGGGUUACAUGGCCCACACAUUGGGU ....................(((((...)))))....((((..((((.((.((((((((.......)))))))).)).))))........(((.((((((....))))))))).)))).. (-24.46 = -23.43 + -1.02)
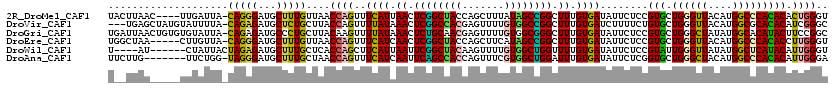
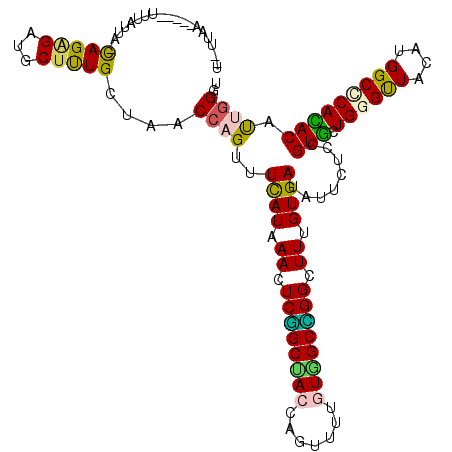
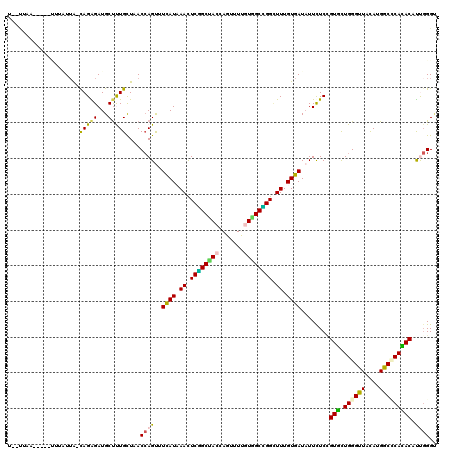
| Location | 12,075,116 – 12,075,219 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -13.48 |
| Energy contribution | -12.57 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
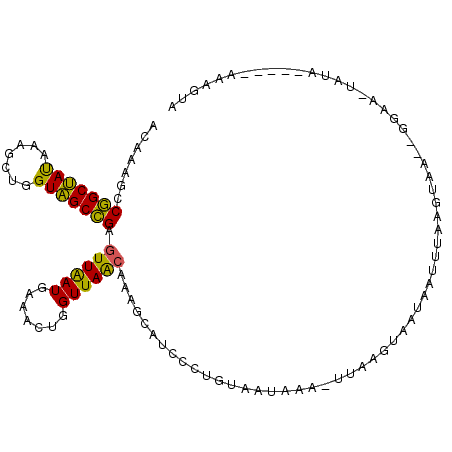
>2R_DroMel_CAF1 12075116 103 + 20766785 ACAAAGCCGGCUAUAAAGCUGGUAGCCGAGUUAAUGAAACUGGUUAACAAAGCAUCCCUGUAAUCAAGUUAAGUAAUAAUUUAAGUGA--GGAAUUAUU-----GAAGUA .....(((((((....)))))))((((.((((.....))))))))......((.((...((((((.(.((((((....)))))).)..--.).))))).-----)).)). ( -19.50) >DroSec_CAF1 70648 108 + 1 ACAAAGCCGGCUAUAAAGCUGGUAGCCGAGUUAAUGAAACUGGUUAACAAAGCAUCCCUGUAAUAAAGUUAAGUAAUAAUUUAAGUGA--GGAAUAAGUAUAAGAAAGGA .....(((((((....)))))))((((.((((.....))))))))...........(((.((...((((((.....))))))...)))--)).................. ( -18.20) >DroSim_CAF1 71518 96 + 1 ACAAAGCCGGCUAUAAAGCUGGUAGCCGAGUUAAUGAAACUGGUUAACAAGGCAUCCCUGUAAUAAAGUUAAGUAAUAAUUUAAGUGA--UA------------AAAGUA (((..(((((((....))))))).(((..((((((.......))))))..))).....)))......((((..(((....)))..)))--).------------...... ( -21.50) >DroEre_CAF1 70830 101 + 1 ACAAAGCCGGCUAUGAAGCUGGUAGCCGAGUUGAUGAAACUGGUUAACAAAGCAUCCCUGUAACAAG-UUAGCCAAGGAUUUAAUUAG--GGAC-UUUA-----GAAGAA .....(((((((....)))))))((((.((((.....))))))))...((((..((((((....(((-((.......)))))...)))--))))-))).-----...... ( -25.00) >DroYak_CAF1 72554 100 + 1 ACAAAACCGGCUAUAAAGCUGGUAGCCGAGUUGAUGAAACUGGUUAACAAAGCGUCCCUGUAAUAAA---GGAUAAGCAUUUGAGUAA--GUUCGUAUA-----GAAGUA .....(((((((....)))))))((((.((((.....))))))))..((((((...(((.......)---))....)).))))....(--.(((.....-----))).). ( -23.30) >DroAna_CAF1 66414 101 + 1 ACAAAUCCAGCCACGAAACUGGUGGCUGAAUUGAUGAAACUGGUUAGCAAAGCAUCCCUACCAGAA---CAAGAAAUAAUGUAAUCAUUUCCAA-UUUA-----AAAGUA .......(((((((.......)))))))(((((((((..(((((...............))))).(---((........)))..))))...)))-))..-----...... ( -19.86) >consensus ACAAAGCCGGCUAUAAAGCUGGUAGCCGAGUUAAUGAAACUGGUUAACAAAGCAUCCCUGUAAUAAA_UUAAGUAAUAAUUUAAGUAA__GGAA_UAUA_____AAAGUA .......(((((((.......))))))).((((((.......)))))).............................................................. (-13.48 = -12.57 + -0.91)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:36 2006