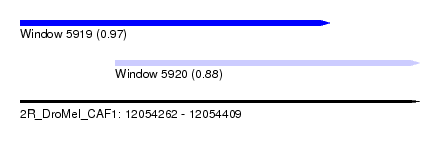
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,054,262 – 12,054,409 |
| Length | 147 |
| Max. P | 0.972977 |

| Location | 12,054,262 – 12,054,376 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
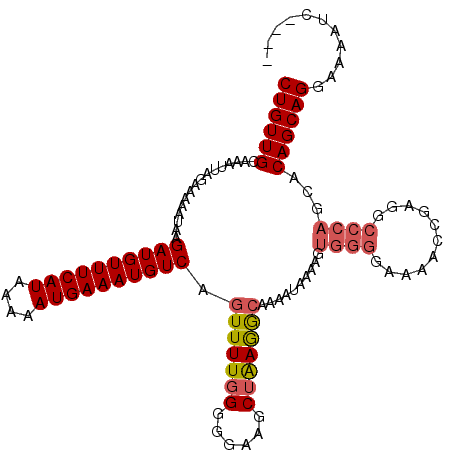
>2R_DroMel_CAF1 12054262 114 + 20766785 -----UAUAAACACAAAAUUGCAGAAAAUGGCGACACCUGCUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAA-AAUGAAAUGUCAGUUUUGGGGGAAGCUAAGGCGAAAUAAAAGUG -----......(((....(((((....((((((.....))))))))))).............((((((((((...-.)))))))))).(((((((......))))))).........))) ( -23.70) >DroVir_CAF1 81246 96 + 1 CGUGUUAUAA--ACAAAAUGGCACAAAAUGGCGACACCUGCUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAAAAAUGAAAUGUCAGA---------------GGCAAAAU------- .(((((((..--.....)))))))......((((((.....))))))...............((((((((((.....))))))))))...---------------........------- ( -21.80) >DroPse_CAF1 109184 110 + 1 -----UAUAAACAGAAAAUUGCAGAAAAUGGCGACACCUGCUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAA-AAUGAAAUGUCAGCUCAAGGGG-CGAAAAGGCGAGA--A-AGGC -----.............((((........((((((.....))))))...............((((((((((...-.)))))))))).(((.....))-)......))))..--.-.... ( -22.70) >DroGri_CAF1 69803 96 + 1 UGUGUUAUAA--ACAAAAUGGCACAAAAUGGCGACACCUGCUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAAAAAUGAAAUGUCAGA---------------GGCGGAAC------- ((((((((..--.....)))))))).....((((((.....))))))...............((((((((((.....))))))))))...---------------........------- ( -22.20) >DroAna_CAF1 51155 114 + 1 -----UAUAAACACAAAAUUGCAGAAAAUGGCGACACCUGCUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAA-AAUGAAAUGUCAGUUUUUGGGGGAGCUAAAAAGGAAGAAAACAA -----.......................((((..(.(((((....)).......((((((..((((((((((...-.)))))))))).))))))))))..))))................ ( -20.60) >DroPer_CAF1 106897 110 + 1 -----UAUAAACAGAAAAUUGCAGAAAAUGGCGACACCUGCUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAA-AAUGAAAUGUCAGCUCAAGGGG-CGAAAAGGCGAGA--A-AGGC -----.............((((........((((((.....))))))...............((((((((((...-.)))))))))).(((.....))-)......))))..--.-.... ( -22.70) >consensus _____UAUAAACACAAAAUUGCAGAAAAUGGCGACACCUGCUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAA_AAUGAAAUGUCAGAU___GGGG__G__AAGGCGAAA__A_AG__ ..................((((........((((((.....))))))...............((((((((((.....))))))))))...................)))).......... (-16.95 = -17.40 + 0.45)
| Location | 12,054,297 – 12,054,409 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -21.45 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12054297 112 + 20766785 CUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAAAAUGAAAUGUCAGUUUUGGGGGAAGCUAAGGCGAAAUAAAAGUGGGGAAAACCGAGGCCCAGCACAGCAGGAAAAUC---- ((((((................((((((((((....))))))))))....(((((....((....))..........(((......)))...)))))..)))))).......---- ( -26.20) >DroSec_CAF1 55156 112 + 1 CUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAAAAUGAAAUGUCUGUUUUGGGGGAAGCUAAGGCAAAAUAAAAGUGGGGAAAACCGAGGCCCAGCACAGCAGGAAAAUC---- ((((((.........((((((.((((((((((....)))))))))))))))).(((...((....))..........(((......)))...)))....)))))).......---- ( -27.00) >DroSim_CAF1 55744 112 + 1 CUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAAAAUGAAAUGUCAGUUUUGGGGGAAGCUGAGGCAAAAUAAAAGUGGGGAAAACCGAGGCCCAGCACAGCAGGAAAAUC---- ((((((................((((((((((....)))))))))).............((((.(((..........(((......)))..))))))).)))))).......---- ( -29.30) >DroEre_CAF1 55390 112 + 1 CUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAAAAUGAAAUGUCAGUUUUGGGGGAAGCUAAGGCGAAAUAAAAGUGGGGAAAGCCGAGGCCCAGCACAGCAGGGAAAUC---- ((((((................((((((((((....)))))))))).(((((((......))))))).........((.(((..........))).)).)))))).......---- ( -25.90) >DroYak_CAF1 56733 112 + 1 CUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAAAAUGAAAUGUCAGUUUUGGGGGAAGCUAAGGCAAAAUAAAAGUGGGCAAAACCGAGGCCCAGCACAGCAGGAAAAUC---- ((((((................((((((((((....)))))))))).(((((((......)))))))..........(((((.........)))))...)))))).......---- ( -28.80) >DroAna_CAF1 51190 112 + 1 CUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAAAAUGAAAUGUCAGUUUUUGGGGGAGCUAAAAAGGAAGAAAACAAGCAAGACUUCAG----GGCACAGCAGGAGUGGCAACA ..(((((...............((((((((((....))))))))))..((((((..(..(((......((((.............))))..----))).)..))))))..))))). ( -23.52) >consensus CUGUUGCAAAUUAGAAAAAUAAGAUGUUUCAUAAAAAUGAAAUGUCAGUUUUGGGGGAAGCUAAGGCAAAAUAAAAGUGGGGAAAACCGAGGCCCAGCACAGCAGGAAAAUC____ ((((((................((((((((((....)))))))))).(((((((......)))))))..........((((...........))))...))))))........... (-21.45 = -22.03 + 0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:30 2006