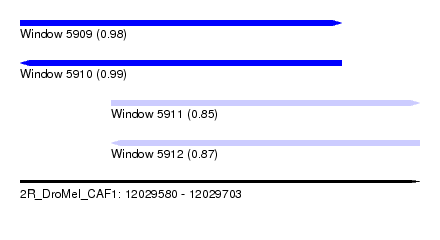
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,029,580 – 12,029,703 |
| Length | 123 |
| Max. P | 0.989428 |

| Location | 12,029,580 – 12,029,679 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -18.54 |
| Energy contribution | -17.98 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12029580 99 + 20766785 UGUUUG---------UGUACGUGUUGGGGCAUACGCG-----CUGUG-----CAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU--GUUGAAAGUUGACAUUG ((((.(---------(((((.((((((.((....)).-----))).)-----))..)))))).......((((.(((..((((((((.....)))))))).--..))).))))))))... ( -28.30) >DroVir_CAF1 32632 103 + 1 UGUGU-------UCUGGUACGUGUCAAAGCGCACACA-----GUGUGCCC---AAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU--GUUGAAAGUUGACAUUG .....-------........(((((..(((((((...-----.((....)---)..)))))))......((((.(((..((((((((.....)))))))).--..))).))))))))).. ( -31.10) >DroGri_CAF1 32928 92 + 1 UGUUUGUCAAAAAGUAGUACGUGUCAAAGC--------------G------------UCAGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU--GUUGAAAGUUGACAUUG ....((((((........((((......))--------------)------------)(((((......)))))(((..((((((((.....)))))))).--..)))...))))))... ( -24.80) >DroWil_CAF1 39602 101 + 1 -----C---------UGUACGUGUCAAAGCAUACGCAUGCAGUAGAG-----AGAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUUGUUUGAAAGUUGACAUUG -----.---------.....(((((........(((((.(......)-----....)))))........((((.(((..((((((((.....)))))))).....))).))))))))).. ( -23.20) >DroMoj_CAF1 39586 97 + 1 UGUAU-------UGUGGUACGUGUCAAA--------------GUGUGCCCCGAGAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU--GUUGAAAGUUGACAUUG .((((-------....))))(((((..(--------------(((..(........)..))))......((((.(((..((((((((.....)))))))).--..))).))))))))).. ( -28.50) >DroPer_CAF1 79083 99 + 1 UGUUUG---------UGUACGUGUCAAAGCAUACGCG-----UUGUG-----CAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCUUUU--GUUGAAAGUUGACAUUG ((((.(---------(((((.((((((.((....)).-----))).)-----))..)))))).......((((.(((..((((((((.....)))))))).--..))).))))))))... ( -25.00) >consensus UGUUUG_________UGUACGUGUCAAAGCAUACGC______GUGUG______AAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU__GUUGAAAGUUGACAUUG ....................(((((((.................................(((......)))..(((..((((((((.....)))))))).....)))...))))))).. (-18.54 = -17.98 + -0.55)

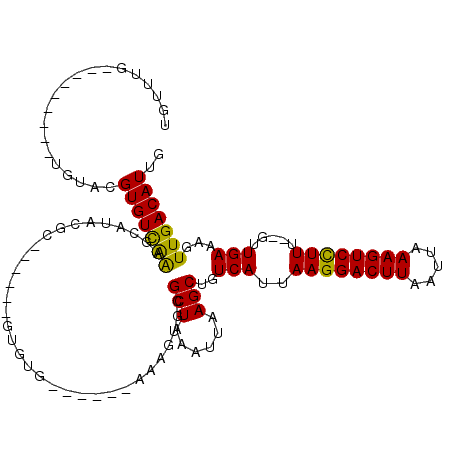
| Location | 12,029,580 – 12,029,679 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -15.33 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12029580 99 - 20766785 CAAUGUCAACUUUCAAC--AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUG-----CACAG-----CGCGUAUGCCCCAACACGUACA---------CAAACA ...(((((.........--.((((((((.....))))))))..)))))..........((((......-----....)-----)))(((((........))))).---------...... ( -21.30) >DroVir_CAF1 32632 103 - 1 CAAUGUCAACUUUCAAC--AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUU---GGGCACAC-----UGUGUGCGCUUUGACACGUACCAGA-------ACACA ...((((((((.(((..--.((((((((.....))))))))..))).))........(((((((..(---((.....)-----)).))))))))))))).........-------..... ( -28.80) >DroGri_CAF1 32928 92 - 1 CAAUGUCAACUUUCAAC--AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCUGA------------C--------------GCUUUGACACGUACUACUUUUUGACAAACA ...((((((...(((..--.((((((((.....))))))))..)))(((((......)))))(------------(--------------(........)))........)))))).... ( -23.00) >DroWil_CAF1 39602 101 - 1 CAAUGUCAACUUUCAAACAAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUCU-----CUCUACUGCAUGCGUAUGCUUUGACACGUACA---------G----- ...((((((...........((((((((.....)))))))).(((.(((((......)))(((.....-----......))).))))).....))))))......---------.----- ( -20.10) >DroMoj_CAF1 39586 97 - 1 CAAUGUCAACUUUCAAC--AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUCUCGGGGCACAC--------------UUUGACACGUACCACA-------AUACA ...((((((...(((..--.((((((((.....))))))))..)))..(((......)))((.((.....))))....--------------.)))))).(((.....-------.))). ( -20.30) >DroPer_CAF1 79083 99 - 1 CAAUGUCAACUUUCAAC--AAAAGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUG-----CACAA-----CGCGUAUGCUUUGACACGUACA---------CAAACA ...((((((........--....(((((.....)))))..........(((......)))(((...((-----(....-----.)))..))).))))))......---------...... ( -16.50) >consensus CAAUGUCAACUUUCAAC__AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUU______CACAA______GCGUAUGCUUUGACACGUACA_________CAAACA ...((((((...........((((((((.....)))))))).......(((......))).................................))))))..................... (-15.33 = -16.00 + 0.67)

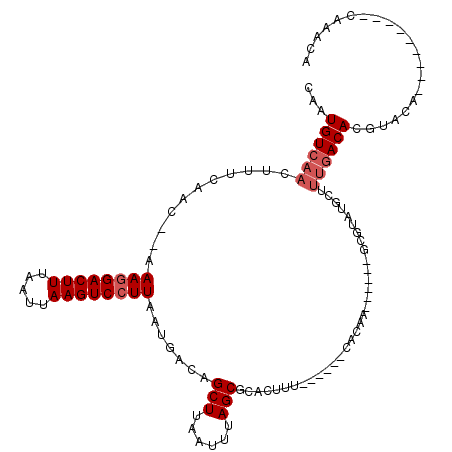
| Location | 12,029,608 – 12,029,703 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -17.11 |
| Energy contribution | -17.09 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12029608 95 + 20766785 --CUGUG-----CAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU--GUUGAAAGUUGACAUUGUGUGAAAGAAACGUU------------UGCG-AAAA---C --...((-----((((((((((.......((((.(((..((((((((.....)))))))).--..))).))))......)))).......)).))------------))))-....---. ( -22.83) >DroVir_CAF1 32662 108 + 1 --GUGUGCCC---AAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU--GUUGAAAGUUGACAUUGUGUAA-AGAAACGUUUUGCUACUGUACUGCG-AAAA---C --(((..(..---...)..))).......((((.(((..((((((((.....)))))))).--..))).))))............-.....(((..(((....)))..)))-....---. ( -25.20) >DroGri_CAF1 32958 98 + 1 ----G------------UCAGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU--GUUGAAAGUUGACAUUGUGUAA-AGAAAACGUUUGCUGCUG--CUGCG-AAAAAUAC ----(------------((((((...........(((..((((((((.....)))))))).--..))).))))))).........-......(((..((....)--).)))-........ ( -22.90) >DroWil_CAF1 39628 100 + 1 AGUAGAG-----AGAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUUUGUUUGAAAGUUGACAUUGUGUAAAGAAAACGUU------------UUGGCGCAA---C .......-----.....(((((((((...((((.(((..((((((((.....)))))))).....))).)))).((.....))...........)------------)))))))).---. ( -25.00) >DroMoj_CAF1 39607 110 + 1 --GUGUGCCCCGAGAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU--GUUGAAAGUUGACAUUGUGUAA-GGAAACAUUUUGCUGCUGUA-UGCG-AAAA---C --((((((..((.(((((((((.......((((.(((..((((((((.....)))))))).--..))).))))......))))..-(....))))))..))..)))-))).-....---. ( -27.82) >DroPer_CAF1 79111 95 + 1 --UUGUG-----CAAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCUUUU--GUUGAAAGUUGACAUUGUGUGAAAGAAACGUU------------UGCG-AAAA---C --...((-----((((((((((.......((((.(((..((((((((.....)))))))).--..))).))))......)))).......)).))------------))))-....---. ( -20.13) >consensus __GUGUG______AAAGUGCGCUAAAUUAAGCUGUCAUUAAGGACUUAAUUAAAGUCCUUU__GUUGAAAGUUGACAUUGUGUAA_AGAAACGUU____________UGCG_AAAA___C .................(((((.......((((.(((..((((((((.....)))))))).....))).))))......))))).................................... (-17.11 = -17.09 + -0.03)

| Location | 12,029,608 – 12,029,703 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -18.62 |
| Consensus MFE | -12.89 |
| Energy contribution | -13.06 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12029608 95 - 20766785 G---UUUU-CGCA------------AACGUUUCUUUCACACAAUGUCAACUUUCAAC--AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUG-----CACAG-- .---....-.(((------------((.((.............(((((.........--.((((((((.....))))))))..)))))(((......)))..))))))-----)....-- ( -19.30) >DroVir_CAF1 32662 108 - 1 G---UUUU-CGCAGUACAGUAGCAAAACGUUUCU-UUACACAAUGUCAACUUUCAAC--AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUU---GGGCACAC-- .---....-.((.(((.((.(((.....))).))-.))).(((.(((.....(((..--.((((((((.....))))))))..)))..(((......)))).)).))---).))....-- ( -21.00) >DroGri_CAF1 32958 98 - 1 GUAUUUUU-CGCAG--CAGCAGCAAACGUUUUCU-UUACACAAUGUCAACUUUCAAC--AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCUGA------------C---- (((.....-((..(--(....))...))......-.))).............(((..--.((((((((.....))))))))..)))(((((......))))).------------.---- ( -18.40) >DroWil_CAF1 39628 100 - 1 G---UUGCGCCAA------------AACGUUUUCUUUACACAAUGUCAACUUUCAAACAAAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUCU-----CUCUACU .---.(((((.((------------(..((.......))....(((((............((((((((.....))))))))..)))))......))).))))).....-----....... ( -18.54) >DroMoj_CAF1 39607 110 - 1 G---UUUU-CGCA-UACAGCAGCAAAAUGUUUCC-UUACACAAUGUCAACUUUCAAC--AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUCUCGGGGCACAC-- .---....-.((.-....)).......(((.(((-(.......(((((.........--.((((((((.....))))))))..)))))(((......))).........)))).))).-- ( -20.60) >DroPer_CAF1 79111 95 - 1 G---UUUU-CGCA------------AACGUUUCUUUCACACAAUGUCAACUUUCAAC--AAAAGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUUG-----CACAA-- .---....-.(((------------((.((.............(((((..(((....--.)))(((((.....))))).....)))))(((......)))..))))))-----)....-- ( -13.90) >consensus G___UUUU_CGCA____________AACGUUUCU_UUACACAAUGUCAACUUUCAAC__AAAGGACUUUAAUUAAGUCCUUAAUGACAGCUUAAUUUAGCGCACUUU______CACAA__ ...........................................(((((............((((((((.....))))))))..)))))(((......))).................... (-12.89 = -13.06 + 0.17)
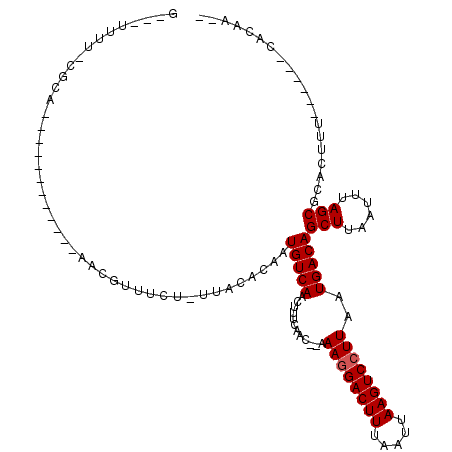
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:22 2006