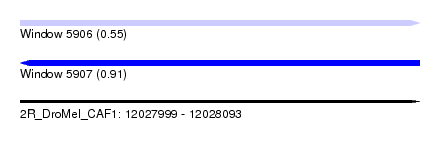
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,027,999 – 12,028,093 |
| Length | 94 |
| Max. P | 0.909961 |

| Location | 12,027,999 – 12,028,093 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -12.95 |
| Energy contribution | -12.04 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12027999 94 + 20766785 ACAUGCAAUUAUG--------------UUUUGUG-----------UGCGAUAACUUGUCAUUAUGCAAAAUAUGCUGCAUUAUUUAUAGCAUAAACACAGGCGCUGACAUUGCAUGUCA (((((((...(((--------------((....(-----------((((((.....))).((((((.(((((........)))))...))))))......)))).)))))))))))).. ( -23.30) >DroVir_CAF1 31546 102 + 1 ACACGCGUUUAUU----UGAUUCGAUGU-AUUUGGCC------------GUAAGUUGUCAUUAUGCAAAAUAUGUUGCAUUAUUUAUAGCAUAAAUACAGGCGCUGACAUUGCAUGUCU ....(((((....----........(((-(((((((.------------.(((((.......((((((......)))))).)))))..)).))))))))))))).(((((...))))). ( -22.10) >DroPse_CAF1 80960 107 + 1 ACAUGCAAUUAUUACACUGC-------UUUUGUGG-----UUUUUUGCUAGAGCUUGUCAUUAUGCAAAAUAUGCUGCAUUAUUUAUGGCAUAAACACAGGCGCUGACGUUGCGUGCCG .((((((((......((..(-------....)..)-----).....(.(((.((((((..((((((.(((((........)))))...))))))..)))))).))).)))))))))... ( -28.80) >DroMoj_CAF1 37878 115 + 1 ACGCGCGUUUAUU----UGCUUCGAUGUUAUUUGGUCGACUCGAGAGUCGAGAGUUGUCAUUAUGCAAAAUAUGUUGCAUUAUUUAUAGCAUAAAUACAGGCGCUGACAUUGCAUGUCC ..((((.(.((((----((..(((((........)))))((((.....)))).(((((....((((((......)))))).....))))).)))))).).)))).(((((...))))). ( -29.70) >DroAna_CAF1 24809 93 + 1 ACAUGCCAUUAUG--------------UU-UGUG-----------UGCGAUAACUUGUCAUUAUGCAAAAUAUGCUGCAUUAUUUAUAGCAUAAACACAGGCGCUGACAUUGCAUGUCA ((((((....(((--------------((-...(-----------((((((.....))).((((((.(((((........)))))...))))))......)))).))))).)))))).. ( -22.70) >DroPer_CAF1 77767 107 + 1 ACAUGCAAUUAUUACACUGC-------UUUUGUGG-----UUUUUCGCUAGAGCUUGUCAUUAUGCAAAAUAUGCUGCAUUAUUUAUGGCAUAAACACAGGCGCUGACGUUGCGUGCCG .(((((((.......((..(-------....)..)-----)....((.(((.((((((..((((((.(((((........)))))...))))))..)))))).))).)))))))))... ( -29.20) >consensus ACAUGCAAUUAUU____UGC_______UUUUGUGG__________UGCGAUAACUUGUCAUUAUGCAAAAUAUGCUGCAUUAUUUAUAGCAUAAACACAGGCGCUGACAUUGCAUGUCA .((((((((......................((...................))..((((...(((....(((((((.(.....).))))))).......))).))))))))))))... (-12.95 = -12.04 + -0.91)

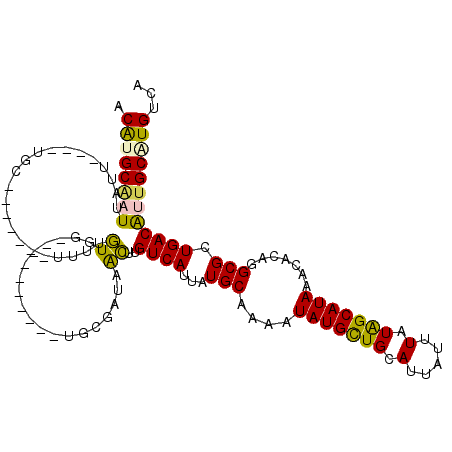
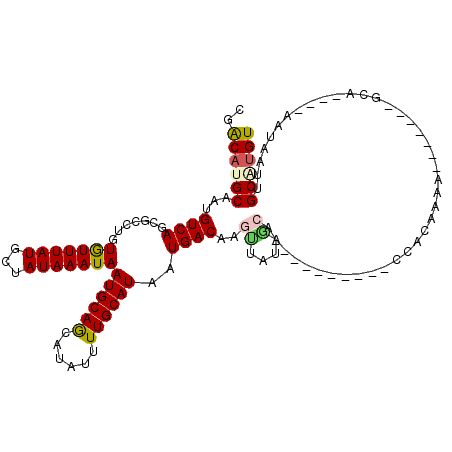
| Location | 12,027,999 – 12,028,093 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -22.29 |
| Consensus MFE | -14.73 |
| Energy contribution | -14.68 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12027999 94 - 20766785 UGACAUGCAAUGUCAGCGCCUGUGUUUAUGCUAUAAAUAAUGCAGCAUAUUUUGCAUAAUGACAAGUUAUCGCA-----------CACAAAA--------------CAUAAUUGCAUGU ..(((((((((.........(((((....(((....((.((((((......)))))).))....)))....)))-----------)).....--------------....))))))))) ( -22.87) >DroVir_CAF1 31546 102 - 1 AGACAUGCAAUGUCAGCGCCUGUAUUUAUGCUAUAAAUAAUGCAACAUAUUUUGCAUAAUGACAACUUAC------------GGCCAAAU-ACAUCGAAUCA----AAUAAACGCGUGU ..((((((..(((((.......(((((((...)))))))((((((......))))))..))))).....(------------((......-...))).....----.......)))))) ( -18.20) >DroPse_CAF1 80960 107 - 1 CGGCACGCAACGUCAGCGCCUGUGUUUAUGCCAUAAAUAAUGCAGCAUAUUUUGCAUAAUGACAAGCUCUAGCAAAAAA-----CCACAAAA-------GCAGUGUAAUAAUUGCAUGU ..(((.((((.(..((.((.(((..((((((...(((((........))))).))))))..))).)).))..)......-----.(((....-------...)))......)))).))) ( -21.00) >DroMoj_CAF1 37878 115 - 1 GGACAUGCAAUGUCAGCGCCUGUAUUUAUGCUAUAAAUAAUGCAACAUAUUUUGCAUAAUGACAACUCUCGACUCUCGAGUCGACCAAAUAACAUCGAAGCA----AAUAAACGCGCGU .(((((...))))).((((...((((..((((.......((((((......))))))...........((((((....))))))..............))))----))))...)))).. ( -27.90) >DroAna_CAF1 24809 93 - 1 UGACAUGCAAUGUCAGCGCCUGUGUUUAUGCUAUAAAUAAUGCAGCAUAUUUUGCAUAAUGACAAGUUAUCGCA-----------CACA-AA--------------CAUAAUGGCAUGU ((((((...))))))..(((((((((((((((.((.....)).)))))....((((((((.....))))).)))-----------...)-))--------------))))..))).... ( -20.80) >DroPer_CAF1 77767 107 - 1 CGGCACGCAACGUCAGCGCCUGUGUUUAUGCCAUAAAUAAUGCAGCAUAUUUUGCAUAAUGACAAGCUCUAGCGAAAAA-----CCACAAAA-------GCAGUGUAAUAAUUGCAUGU ..(((.((((((..((.((.(((..((((((...(((((........))))).))))))..))).)).))..)).....-----.(((....-------...)))......)))).))) ( -23.00) >consensus CGACAUGCAAUGUCAGCGCCUGUGUUUAUGCUAUAAAUAAUGCAGCAUAUUUUGCAUAAUGACAAGUUAUAGCA__________CCACAAAA_______GCA____AAUAAUUGCAUGU ..((((((...((((.......(((((((...)))))))((((((......))))))..))))..((....))........................................)))))) (-14.73 = -14.68 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:17 2006