| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,025,255 – 12,025,347 |
| Length | 92 |
| Max. P | 0.970503 |

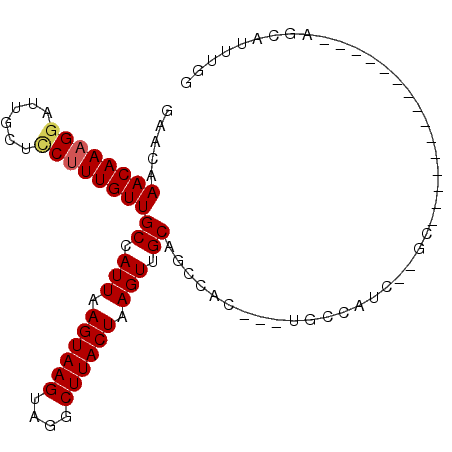
| Location | 12,025,255 – 12,025,347 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.48 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -14.74 |
| Energy contribution | -16.30 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12025255 92 + 20766785 CCAAAUGGCAGAAAUGGCGAGAUGGC------GCA---GCGGCUGCAACUUAGUAAGCCUACUUACUUAAUGGCAACAAAGAAGCAAUCCUUUGUUUGUUC (((....((((.....(((......)------)).---....)))).....((((((....))))))...))).(((((((........)))))))..... ( -21.30) >DroSec_CAF1 26570 98 + 1 CCAAAUGGCAGAAAUGGCGAGAUGGCGAGAUGGCA---GCGGCUGCAACUUAGUAAGCCUACUUACUUAAUGGCAACAAAGGAGCAAUCCUUUGUUUGCUC (((..((.((....)).))...))).(((...(((---(...))))..)))((((((....))))))....((((((((((((....))))))).))))). ( -29.10) >DroEre_CAF1 26427 83 + 1 CCAAAUGCU---------------GCGAGAUGGCA---GUGGCUGCAACUUAGUAAGCCUACUUACUUAAUGGCAACAAAGGAGCAAUCCUUUGUUUGUUC ......(((---------------((......)))---))....((.....((((((....)))))).....))(((((((((....)))))))))..... ( -26.20) >DroYak_CAF1 26386 86 + 1 CCAAAUGCU---------------GCGAGAUGGCAGCCGUGGCUGCAACUUAGUAAGCCUACUUACUUAAUGGCAACAAAGGAGCAAGCCUUUGUUUGUUC (((...(((---------------((......)))))..)))..((.....((((((....)))))).....))((((((((......))))))))..... ( -27.10) >DroMoj_CAF1 32071 71 + 1 UC----GU-----------------------UGCU---GCAGCUGCAACUUAGUAAGCCUACUUACUUAAUGGCAACAAAGGCACAAUUCAUUGUUUGUGC ..----((-----------------------((((---((....)).....((((((....))))))....))))))....((((((........)))))) ( -19.90) >DroAna_CAF1 22288 81 + 1 CUAACUGCU---------------UC--GACGGCA---GUUGCUGCAACUUAGUAAGCCUACUUACUUAAUGGCAACAAAGGAGCAAUCCUUUGUUUGUAC .((((((((---------------..--...))))---))))..((.....((((((....)))))).....))(((((((((....)))))))))..... ( -27.90) >consensus CCAAAUGCU_______________GC__GAUGGCA___GCGGCUGCAACUUAGUAAGCCUACUUACUUAAUGGCAACAAAGGAGCAAUCCUUUGUUUGUUC ................................((.......)).((.....((((((....)))))).....))(((((((((....)))))))))..... (-14.74 = -16.30 + 1.56)

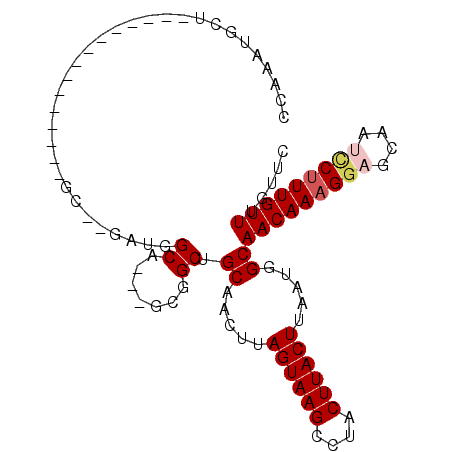
| Location | 12,025,255 – 12,025,347 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.48 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -16.61 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12025255 92 - 20766785 GAACAAACAAAGGAUUGCUUCUUUGUUGCCAUUAAGUAAGUAGGCUUACUAAGUUGCAGCCGC---UGC------GCCAUCUCGCCAUUUCUGCCAUUUGG .....(((((((((....)))))))))((((((.....))).)))...((((((.((((....---.((------(......))).....)))).)))))) ( -21.60) >DroSec_CAF1 26570 98 - 1 GAGCAAACAAAGGAUUGCUCCUUUGUUGCCAUUAAGUAAGUAGGCUUACUAAGUUGCAGCCGC---UGCCAUCUCGCCAUCUCGCCAUUUCUGCCAUUUGG .(((.(((((((((....)))))))))((.(((.((((((....)))))).))).))....))---).........(((....((.......))....))) ( -24.60) >DroEre_CAF1 26427 83 - 1 GAACAAACAAAGGAUUGCUCCUUUGUUGCCAUUAAGUAAGUAGGCUUACUAAGUUGCAGCCAC---UGCCAUCUCGC---------------AGCAUUUGG .....(((((((((....)))))))))((.(((.((((((....)))))).))).))..((((---(((......))---------------))....))) ( -24.10) >DroYak_CAF1 26386 86 - 1 GAACAAACAAAGGCUUGCUCCUUUGUUGCCAUUAAGUAAGUAGGCUUACUAAGUUGCAGCCACGGCUGCCAUCUCGC---------------AGCAUUUGG .....((((((((......))))))))((.(((.((((((....)))))).))).))..(((..(((((......))---------------)))...))) ( -27.70) >DroMoj_CAF1 32071 71 - 1 GCACAAACAAUGAAUUGUGCCUUUGUUGCCAUUAAGUAAGUAGGCUUACUAAGUUGCAGCUGC---AGCA-----------------------AC----GA ((((((........))))))..(((((((.....((((((....))))))....(((....))---))))-----------------------))----)) ( -21.50) >DroAna_CAF1 22288 81 - 1 GUACAAACAAAGGAUUGCUCCUUUGUUGCCAUUAAGUAAGUAGGCUUACUAAGUUGCAGCAAC---UGCCGUC--GA---------------AGCAGUUAG ((...(((((((((....)))))))))((.(((.((((((....)))))).))).)).))(((---(((....--..---------------.)))))).. ( -25.90) >consensus GAACAAACAAAGGAUUGCUCCUUUGUUGCCAUUAAGUAAGUAGGCUUACUAAGUUGCAGCCAC___UGCCAUC__GC_______________AGCAUUUGG .....((((((((......))))))))((.(((.((((((....)))))).))).))............................................ (-16.61 = -16.83 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:14 2006