
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,009,172 – 12,009,273 |
| Length | 101 |
| Max. P | 0.749509 |

| Location | 12,009,172 – 12,009,273 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -20.16 |
| Energy contribution | -21.63 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
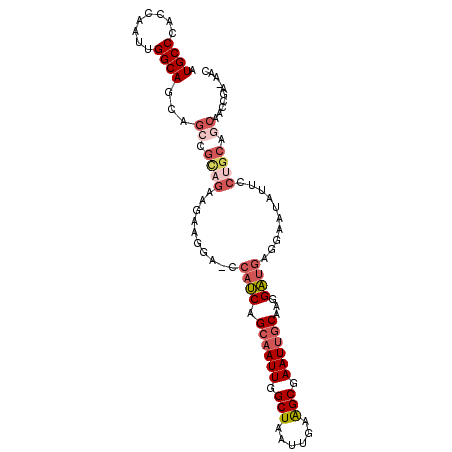
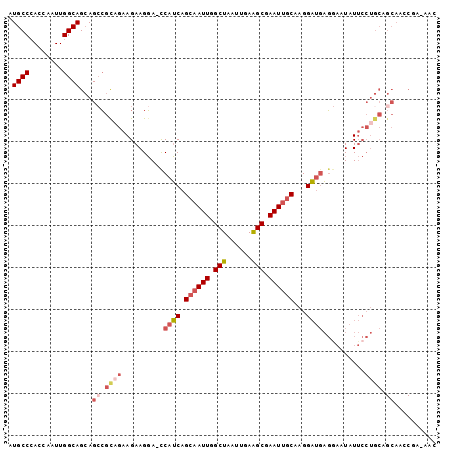
>2R_DroMel_CAF1 12009172 101 + 20766785 AUGCCCACCAAUUGGCAGCAGCCGCAGAAGAAGGAAUCAUCAGCAAUUGGCUAAUUGAAGCGAAUUGCAAGGAUGAGGAAUAUUCCUGCAGCAACCGA-AAC .((((........))))...((.((((..(((....(((((.((((((.(((......))).))))))...)))))......))))))).))......-... ( -31.30) >DroSec_CAF1 10490 101 + 1 AUGCCCACCAAUUGGCAGCAGGCGCCGAAGAAGGA-CCAUCAGCAAUUGGCUAAUUGAAGCGAAUUGCAAGGAUGAGAAAUAUUCCUGCAGCAACCGACAAC .((((........))))(((((..((......)).-.((((.((((((.(((......))).))))))...)))).........)))))............. ( -30.10) >DroSim_CAF1 10515 101 + 1 AUGCCCACCAAUUGGCAGCAGCCGCAGAAGAAGGA-CCAUCAGCAAUUGGCUAAUUGAAGCGAAUUGCAAGGAUGAGGAAUAUUCCUGCAGCAACCGACAAC .((((........))))...((.((((..(((...-(((((.((((((.(((......))).))))))...)))).).....))))))).)).......... ( -29.30) >DroEre_CAF1 8233 100 + 1 AUGCCCACGAAUUGGCAGCAGCCGCAGAAGAAGGA-CCAUCAGCAAUUGGCUAAUUGAAGCGAAUUGCAAGGAUGAGGAAUAUUCCUGCAGCAAACCG-AAC .((((........))))...((.((((..(((...-(((((.((((((.(((......))).))))))...)))).).....))))))).))......-... ( -29.30) >DroYak_CAF1 10313 100 + 1 AUGCCCACCAAUUGGCAGCAGCCGUAGAAGAAGGA-CCAUCAGCAAUUGGCUAAUUGAGGCCAAUUGCAAGGAUGAGGAAUAUUCCUGCAGCAAGCCA-AAC ...........(((((.((.((.............-.((((.((((((((((......))))))))))...))))((((....)))))).))..))))-).. ( -32.70) >DroAna_CAF1 6433 87 + 1 AUGCCGAACAAUUGGCAGGAGCAGUACAAGAAGUAUCCAACAGCAAUUGGCUAAUUGAAGCGAAU-ACGAGGUCUUUGUAUAU--------------ACAAA .((((((....))))))......((((((((.((((.(..(((...)))(((......)))).))-))....)).))))))..--------------..... ( -15.60) >consensus AUGCCCACCAAUUGGCAGCAGCCGCAGAAGAAGGA_CCAUCAGCAAUUGGCUAAUUGAAGCGAAUUGCAAGGAUGAGGAAUAUUCCUGCAGCAACCGA_AAC .((((........))))...((.((((..........((((.((((((.(((......))).))))))...))))..........)))).)).......... (-20.16 = -21.63 + 1.48)

| Location | 12,009,172 – 12,009,273 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -23.35 |
| Energy contribution | -25.36 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
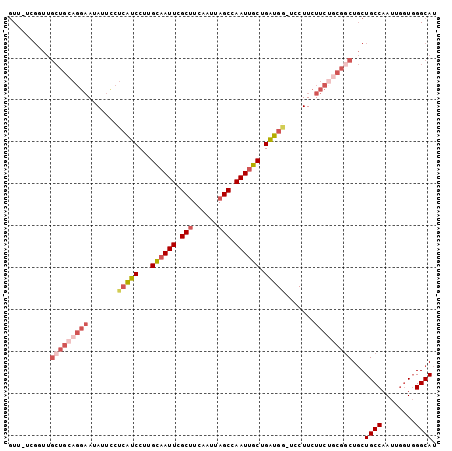
>2R_DroMel_CAF1 12009172 101 - 20766785 GUU-UCGGUUGCUGCAGGAAUAUUCCUCAUCCUUGCAAUUCGCUUCAAUUAGCCAAUUGCUGAUGAUUCCUUCUUCUGCGGCUGCUGCCAAUUGGUGGGCAU ...-..(((.(((((((((.......(((((...((((((.(((......))).)))))).))))).......))))))))).)))(((........))).. ( -35.44) >DroSec_CAF1 10490 101 - 1 GUUGUCGGUUGCUGCAGGAAUAUUUCUCAUCCUUGCAAUUCGCUUCAAUUAGCCAAUUGCUGAUGG-UCCUUCUUCGGCGCCUGCUGCCAAUUGGUGGGCAU .(..((((((((.(((((.........((((...((((((.(((......))).)))))).))))(-((.......))).))))).).)))))))..).... ( -31.50) >DroSim_CAF1 10515 101 - 1 GUUGUCGGUUGCUGCAGGAAUAUUCCUCAUCCUUGCAAUUCGCUUCAAUUAGCCAAUUGCUGAUGG-UCCUUCUUCUGCGGCUGCUGCCAAUUGGUGGGCAU ..(((((((.(((((((((.......(((((...((((((.(((......))).)))))).)))))-......))))))))).)))(((....))).)))). ( -35.82) >DroEre_CAF1 8233 100 - 1 GUU-CGGUUUGCUGCAGGAAUAUUCCUCAUCCUUGCAAUUCGCUUCAAUUAGCCAAUUGCUGAUGG-UCCUUCUUCUGCGGCUGCUGCCAAUUCGUGGGCAU ...-.(((..(((((((((.......(((((...((((((.(((......))).)))))).)))))-......))))))))).)))(((........))).. ( -33.22) >DroYak_CAF1 10313 100 - 1 GUU-UGGCUUGCUGCAGGAAUAUUCCUCAUCCUUGCAAUUGGCCUCAAUUAGCCAAUUGCUGAUGG-UCCUUCUUCUACGGCUGCUGCCAAUUGGUGGGCAU ...-..(((..(((.((((.......(((((...(((((((((........))))))))).)))))-)))).)......(((....)))....))..))).. ( -34.11) >DroAna_CAF1 6433 87 - 1 UUUGU--------------AUAUACAAAGACCUCGU-AUUCGCUUCAAUUAGCCAAUUGCUGUUGGAUACUUCUUGUACUGCUCCUGCCAAUUGUUCGGCAU ...((--------------(..(((((.((....((-((((((..(((((....)))))..).))))))).))))))).)))...((((........)))). ( -15.70) >consensus GUU_UCGGUUGCUGCAGGAAUAUUCCUCAUCCUUGCAAUUCGCUUCAAUUAGCCAAUUGCUGAUGG_UCCUUCUUCUGCGGCUGCUGCCAAUUGGUGGGCAU ..........(((((((((.......(((((...((((((.(((......))).)))))).))))).......)))))))))...((((........)))). (-23.35 = -25.36 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:05 2006