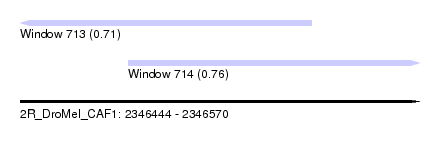
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,346,444 – 2,346,570 |
| Length | 126 |
| Max. P | 0.755751 |

| Location | 2,346,444 – 2,346,536 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -24.29 |
| Energy contribution | -24.02 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
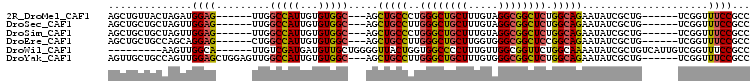
>2R_DroMel_CAF1 2346444 92 - 20766785 AGCUGUUACUAGAUGGAG------UUGGCCAUUGUGUGGC---AGCUGCCCUGGGCUGCUUUGUAGGCGGCUCUGGCAGAAUAUCGCUG------UCGGUUUCCGCC (((((((((..(((((..------....)))))..)))))---))))(((...))).........(((((..(((((((.......)))------))))...))))) ( -38.20) >DroSec_CAF1 1996 92 - 1 AGCUGCUGCUAGUUGGAG------UUGGCCAUUGUGUGGC---AGCUGCCUUGGGCUGCUUUGUAGGCGGCUCUGGCAGAAUAUCGCUG------UCGGUUUCCGCC ((((((..(....(((..------....)))....)..))---))))(((...))).........(((((..(((((((.......)))------))))...))))) ( -37.70) >DroSim_CAF1 2014 92 - 1 AGCUGCUGCUAGUUGGAG------UUGGCCAUUGUGUGGC---AGCUGCCCUGGGCUGCUUUGUAGGCGGCUCUGGCAGAAUAUCGCUG------UCGGUUUCCGCC ((((((..(....(((..------....)))....)..))---))))(((...))).........(((((..(((((((.......)))------))))...))))) ( -37.70) >DroEre_CAF1 1882 92 - 1 AGCUGCUGCCAGCAGGAG------CUGGCCAUUGUGUGGC---AGCUGCCUUGGGCUGCUUGGUGGGCGGCUCCGGCAGAAUAUCGCUG------UCGGUUUCCGCC .((((....)))).((((------(((.(((((....(((---((((......))))))).))))).)))))))(((.(((.((((...------.))))))).))) ( -43.90) >DroWil_CAF1 3131 92 - 1 ---------AAGUUGGCA------UUGUCGAUGAUGUUGCUGGGGUUACUGGUGGCCCCUUUGUUGGCGGUUCUGGCAAAAUAUCGCUGUCAUUGUCGGUUUCCGCC ---------.....((((------..(((...)))..))))(((((((....)))))))......(((((..(((((((.............)))))))...))))) ( -30.72) >DroYak_CAF1 1873 98 - 1 AGUUGCUGCCAGUUGGAGCUGGAGUUGGCCAUUGUGUGGC---AGCUGCCUUGGGCUGCUUUGUGGGCGGCUCUGGCAGAAUAUCGCUG------UCGGUUUCCGCC .(..((((.((((....((..((((((.((((.....(((---((((......)))))))..)))).))))))..)).(.....)))))------.))))..).... ( -40.40) >consensus AGCUGCUGCUAGUUGGAG______UUGGCCAUUGUGUGGC___AGCUGCCUUGGGCUGCUUUGUAGGCGGCUCUGGCAGAAUAUCGCUG______UCGGUUUCCGCC ..............((((.........(((((...))))).....(((((..((((((((.....)))))))).))))).....................))))... (-24.29 = -24.02 + -0.27)

| Location | 2,346,478 – 2,346,570 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.57 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.41 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2346478 92 + 20766785 CCUACAAAGCAGCCCAGGGCAGCUGCCACACAAUGGCCAA------CUCCAUCUAGUAACAGCUCGGCCAGGGCGUUGAUGACGAGCAACGUGGCAUC .((((......((((.((.((((((..((...((((....------..))))...))..))))).).)).))))((((........)))))))).... ( -29.70) >DroSec_CAF1 2030 92 + 1 CCUACAAAGCAGCCCAAGGCAGCUGCCACACAAUGGCCAA------CUCCAACUAGCAGCAGCUCGGCCAGGGCGUUGAUGACGAGCAACGUGGCAUC .((((......((((..(((((((((.......(((....------..))).......))))))..))).))))((((........)))))))).... ( -29.24) >DroSim_CAF1 2048 92 + 1 CCUACAAAGCAGCCCAGGGCAGCUGCCACACAAUGGCCAA------CUCCAACUAGCAGCAGCUCGGCCAGGGCGUUGAUGACGAGCAACGUGGCAUC .((((......((((.((.(((((((.......(((....------..))).......)))))).).)).))))((((........)))))))).... ( -30.84) >DroEre_CAF1 1916 92 + 1 CCCACCAAGCAGCCCAAGGCAGCUGCCACACAAUGGCCAG------CUCCUGCUGGCAGCAGCUCGGCCAGGGCGUUGAUGGCGAGCAACGUGGCAUC .((((......((((..(((((((((((.....))(((((------(....)))))).))))))..))).))))((((........)))))))).... ( -40.70) >DroYak_CAF1 1907 98 + 1 CCCACAAAGCAGCCCAAGGCAGCUGCCACACAAUGGCCAACUCCAGCUCCAACUGGCAGCAACUCGGCCAGGGCGUUGAUGACGGGCAUGGUGGCAUC .......(((.(((...))).)))(((((...(((.((..(..((((.((..(((((.........))))))).))))..)..)).))).)))))... ( -36.70) >consensus CCUACAAAGCAGCCCAAGGCAGCUGCCACACAAUGGCCAA______CUCCAACUAGCAGCAGCUCGGCCAGGGCGUUGAUGACGAGCAACGUGGCAUC .((((......((((..(((....)))......(((((...........................)))))))))((((........)))))))).... (-26.25 = -26.41 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:26 2006