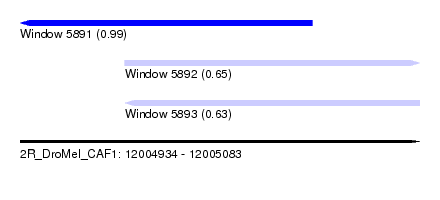
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,004,934 – 12,005,083 |
| Length | 149 |
| Max. P | 0.986941 |

| Location | 12,004,934 – 12,005,043 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -43.64 |
| Consensus MFE | -27.28 |
| Energy contribution | -28.25 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12004934 109 - 20766785 CAUUGCCAUGGUU--AUGUGUGCGACUGUAUGUAGCCA------UGGCGGUGAAUCUGGUUCCGGAUUGUGGAUGUGGCCAAAGGGUAUGGGCAUGGGCGUGUUGGCGACAUUACGC (((((((((((((--(((((........))))))))))------))))))))((((((....))))))((((((((.(((((.(.((..........)).).))))).))))).))) ( -45.20) >DroSec_CAF1 6249 103 - 1 CAUUGCCAUGGUU--AUAUGUUCGACUGUAUGUAGCCA------UGGCGGUGAAUCUGGAUCCGGAUUGUGGAUGUGGCCAAAGGGUAUG------GGCGUGUUGGCGACAUUACGC (((((((((((((--(((((........))))))))))------))))))))((((((....))))))((((((((.(((((.(.((...------.)).).))))).))))).))) ( -47.10) >DroSim_CAF1 6024 103 - 1 CAUUGCCAUGGUU--AUAUGUUCGACUGUAUGUAGCCA------UGGCGGUGAAUCUGGAUCCGGAUUGUGGAUGUGGCCAAAGGGUAUG------GGCGUGUUGGCGACAUUACGC (((((((((((((--(((((........))))))))))------))))))))((((((....))))))((((((((.(((((.(.((...------.)).).))))).))))).))) ( -47.10) >DroEre_CAF1 3986 110 - 1 CAUGGUUAUACGUACAUAUGUGCGACUUUAUGUAGCCAUGGCCAUGGCGGUGAAUCUGGAUACGGAUUGUGGAUGUGGCCAA-GGGUAUG------GGCGUGUUGGCGACAUUACGC (((((((((((((((....)))))......))))))))))((....))....((((((....))))))((((((((.(((((-.(((...------.)).).))))).))))).))) ( -39.00) >DroYak_CAF1 6076 98 - 1 CAUGGUACUACUCGUAUACGAG------UAUGUAGCCA------UGGCGGUGAAUCUGGAUCCGGAUUGUGGAUGUGGCCAA-GGGUAUG------GGCGUGUUGGCGACAUUACGC ((((((((((((((....))))------)).)).))))------))(((...((((((....))))))...(((((.(((((-.(((...------.)).).))))).))))).))) ( -39.80) >consensus CAUUGCCAUGGUU__AUAUGUGCGACUGUAUGUAGCCA______UGGCGGUGAAUCUGGAUCCGGAUUGUGGAUGUGGCCAAAGGGUAUG______GGCGUGUUGGCGACAUUACGC ((((((((........(((((((....)))))))..........))))))))((((((....))))))((((((((.(((((.(.((..........)).).))))).))))).))) (-27.28 = -28.25 + 0.97)

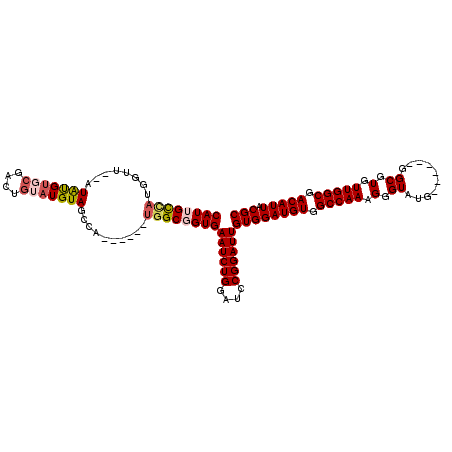
| Location | 12,004,973 – 12,005,083 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.66 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -7.13 |
| Energy contribution | -8.47 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12004973 110 + 20766785 CCACAUCCACAAUCCGGAACCAGAUUCACCGCCA------UGGCUACAUACAGUCGCACACAU--AACCAUGGCAAU-GGCAAUGGCCAUGUCCCACCUCGCCAUCCUCAACAUCCUUC ...............(((............((((------(((....................--..)))))))..(-((..(((((...(.....)...)))))..)))...)))... ( -20.65) >DroSec_CAF1 6282 107 + 1 CCACAUCCACAAUCCGGAUCCAGAUUCACCGCCA------UGGCUACAUACAGUCGAACAUAU--AACCAUGGCAAU-GGCAAUGGCCAUGUCC---CUCGCCAUCCUCAACAUCCUUC ...............((((...((......((((------(((....................--..))))))).((-(((....)))))....---.))...))))............ ( -21.95) >DroSim_CAF1 6057 107 + 1 CCACAUCCACAAUCCGGAUCCAGAUUCACCGCCA------UGGCUACAUACAGUCGAACAUAU--AACCAUGGCAAU-GGCAAUGGCCAUGUCC---CUCGCCAUCCUCAACAUCCUUC ...............((((...((......((((------(((....................--..))))))).((-(((....)))))....---.))...))))............ ( -21.95) >DroEre_CAF1 4018 111 + 1 CCACAUCCACAAUCCGUAUCCAGAUUCACCGCCAUGGCCAUGGCUACAUAAAGUCGCACAUAUGUACGUAUAACCAU-GGCAAUGGCCACGUCC---CUCGC----CUCAACAUCCUUC ..............................(((((.((((((((((((((..........)))))).).....))))-))).))))).......---.....----............. ( -21.30) >DroYak_CAF1 6108 99 + 1 CCACAUCCACAAUCCGGAUCCAGAUUCACCGCCA------UGGCUACAUA------CUCGUAUACGAGUAGUACCAU-GGCAAUGGCCAUGUCC---CUUGC----CUCAACAUCCUUC ...............((((...((......((((------(((.(((.((------((((....)))))))))))))-)))...(((.......---...))----)))...))))... ( -30.00) >DroAna_CAF1 2404 97 + 1 CCACAUCCUCAUU-CACAUCCACUUGCACAACCA------------UAUCCUGAUUCACAUAU--AACCAUGGCAGCGGGCAAUGGCCAUGUCC---AUGCC----CUCAGCAUCCUUC .............-..........(((.......------------....(((.....(((..--....))).))).(((((.(((......))---)))))----)...)))...... ( -15.70) >consensus CCACAUCCACAAUCCGGAUCCAGAUUCACCGCCA______UGGCUACAUACAGUCGCACAUAU__AACCAUGGCAAU_GGCAAUGGCCAUGUCC___CUCGC____CUCAACAUCCUUC ...............((((...........((((......))))..................................(((....)))........................))))... ( -7.13 = -8.47 + 1.33)

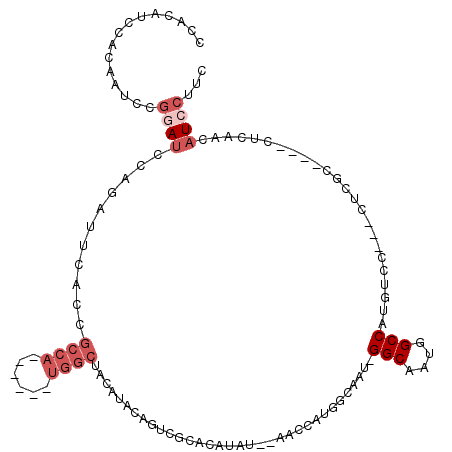
| Location | 12,004,973 – 12,005,083 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.66 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -11.79 |
| Energy contribution | -11.68 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
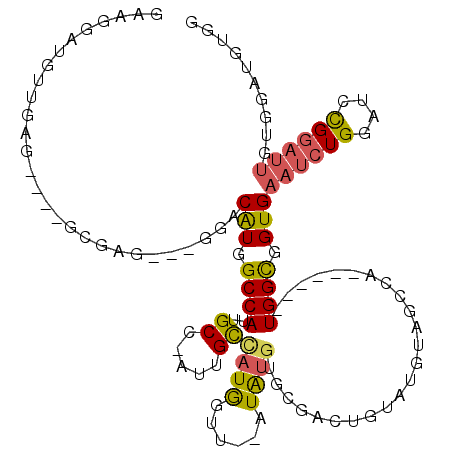
>2R_DroMel_CAF1 12004973 110 - 20766785 GAAGGAUGUUGAGGAUGGCGAGGUGGGACAUGGCCAUUGCC-AUUGCCAUGGUU--AUGUGUGCGACUGUAUGUAGCCA------UGGCGGUGAAUCUGGUUCCGGAUUGUGGAUGUGG .....(((((.(.(((((((((((.(....).))).)))))-)))(((((((((--(((((........))))))))))------))))....((((((....)))))).).))))).. ( -40.10) >DroSec_CAF1 6282 107 - 1 GAAGGAUGUUGAGGAUGGCGAG---GGACAUGGCCAUUGCC-AUUGCCAUGGUU--AUAUGUUCGACUGUAUGUAGCCA------UGGCGGUGAAUCUGGAUCCGGAUUGUGGAUGUGG ...((((.(.((.((((((.(.---.....).))))))..(-((((((((((((--(((((........))))))))))------))))))))..)).).))))............... ( -40.70) >DroSim_CAF1 6057 107 - 1 GAAGGAUGUUGAGGAUGGCGAG---GGACAUGGCCAUUGCC-AUUGCCAUGGUU--AUAUGUUCGACUGUAUGUAGCCA------UGGCGGUGAAUCUGGAUCCGGAUUGUGGAUGUGG ...((((.(.((.((((((.(.---.....).))))))..(-((((((((((((--(((((........))))))))))------))))))))..)).).))))............... ( -40.70) >DroEre_CAF1 4018 111 - 1 GAAGGAUGUUGAG----GCGAG---GGACGUGGCCAUUGCC-AUGGUUAUACGUACAUAUGUGCGACUUUAUGUAGCCAUGGCCAUGGCGGUGAAUCUGGAUACGGAUUGUGGAUGUGG .....(((((.(.----.....---...(((.(((((.(((-((((((((((((((....)))))......)))))))))))).))))).)))((((((....)))))).).))))).. ( -39.00) >DroYak_CAF1 6108 99 - 1 GAAGGAUGUUGAG----GCAAG---GGACAUGGCCAUUGCC-AUGGUACUACUCGUAUACGAG------UAUGUAGCCA------UGGCGGUGAAUCUGGAUCCGGAUUGUGGAUGUGG ......(((....----)))..---..((((...(((((((-(((((((((((((....))))------)).)).))))------))))))))((((((....))))))....)))).. ( -37.00) >DroAna_CAF1 2404 97 - 1 GAAGGAUGCUGAG----GGCAU---GGACAUGGCCAUUGCCCGCUGCCAUGGUU--AUAUGUGAAUCAGGAUA------------UGGUUGUGCAAGUGGAUGUG-AAUGAGGAUGUGG ...(.((.((.((----(((((---((......))).)))))(((..(((..((--((((.(......).)))------------)))..)))..))).......-..).)).)).).. ( -22.90) >consensus GAAGGAUGUUGAG____GCGAG___GGACAUGGCCAUUGCC_AUUGCCAUGGUU__AUAUGUGCGACUGUAUGUAGCCA______UGGCGGUGAAUCUGGAUCCGGAUUGUGGAUGUGG ............................(((.((((..((.....))((((......))))........................)))).)))((((((....)))))).......... (-11.79 = -11.68 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:03 2006