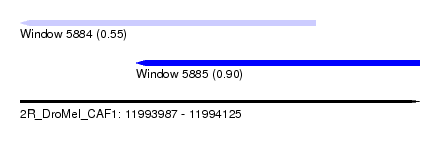
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,993,987 – 11,994,125 |
| Length | 138 |
| Max. P | 0.901329 |

| Location | 11,993,987 – 11,994,089 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -15.02 |
| Energy contribution | -14.85 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
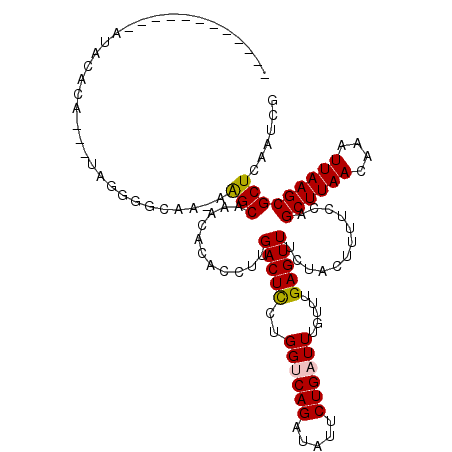
>2R_DroMel_CAF1 11993987 102 - 20766785 ------------AUGGACA---UAGGUAGAA-AGGCAAAUACGCCUUGACUCUCGGUCAGAUAUUCUGAUUUGUUUGAGUUUCUACUUUUCGAGCUUAACAAAUUAAGCGCUCAAUCU ------------.((..(.---.((((((((-((((......)))))(((((.(((((((.....)))).)))...)))))))))))).....((((((....)))))))..)).... ( -28.40) >DroVir_CAF1 48318 102 - 1 AC------------ACAAA---CAAGGGCCA-AAGCAAACACACCUUGACUUCUGGUCAGAUAUUCUGAUUUGUUUGAGUUUCUACUUUUGCAGCUUAACAAAUUAAGCGCUCAAUCG ..------------.....---...((((..-..(((((........((((((..(((((.....)))))..)...)))))......))))).((((((....))))))))))..... ( -22.54) >DroGri_CAF1 46528 115 - 1 ACACACUCACUAAUACAUA---CAGGGGCCAAAAGCAAACACACCUUGACUUCUGGCCAGAUAUUCUGAUUUGUUUGAGUUUCUACUUUUGUAGCUUAACAAAUUAAGCGCUCAAUCG ...................---(((((((((.((((((.......))).))).)))))......))))....(.((((((...(((....)))((((((....)))))))))))).). ( -24.30) >DroEre_CAF1 45355 105 - 1 ------------AUGGACAUUGUAGGUAGAA-AGGCAAAUACGCCUUGACUCUCGGCCAGAUAUUCUGAUUUGUUUGAGUUUCUACUUUUCCAGCUUAACAAAUUAAGCGCUCAAUCU ------------.((((......((((((((-(..((((((.(((.........)))(((.....)))...))))))..))))))))).))))((((((....))))))......... ( -27.30) >DroAna_CAF1 48046 106 - 1 --------AUGUAUGCAUG---UAGGUGAAG-AGGCAAACACACCUUGACUCUUGGUCAGAUAUUCUGAUUUGUUUGAGUUUCUACUUUUCGAGCUUAACAAAUUAAGCGCUCAAUCU --------.((...((..(---((((....(-(((........))))(((((..((((((.....)))))).....)))))))))).......((((((....)))))))).)).... ( -23.70) >DroMoj_CAF1 55832 102 - 1 AC------------AGAGA---UGAGGGCCA-AAGCAAACACACCUUGACUUCUGGUCAGAUAUUCUGAUUUGUUUGAGUUUCUACUUUUGCAGCUUAACAAAUUAAGCGCUCAAUCG ..------------...((---(..((((..-..(((((........((((((..(((((.....)))))..)...)))))......))))).((((((....)))))))))).))). ( -24.24) >consensus ____________AUACACA___UAGGGGCAA_AAGCAAACACACCUUGACUCCUGGUCAGAUAUUCUGAUUUGUUUGAGUUUCUACUUUUCCAGCUUAACAAAUUAAGCGCUCAAUCG .................................(((...........(((((..((((((.....)))))).....)))))............((((((....)))))))))...... (-15.02 = -14.85 + -0.17)

| Location | 11,994,027 – 11,994,125 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.24 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -14.49 |
| Energy contribution | -14.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
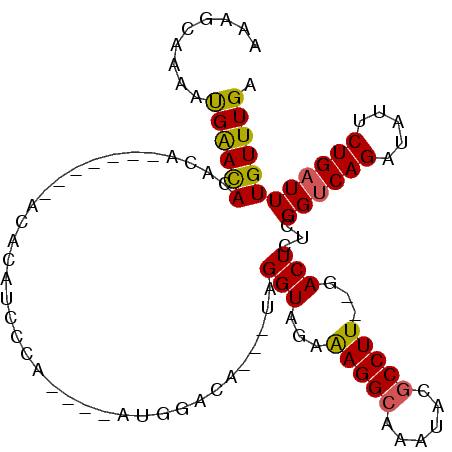
>2R_DroMel_CAF1 11994027 98 - 20766785 AAAGCAAAAUGAAUACACACACACACACACAGCCCA----AUGGACA---UAGGUAGAAAGGCAAAUACGCCUU--GACUCUCGGUCAGAUAUUCUGAUUUGUUUGA .(((((((..((((((.............((.....----.))(((.---.((((...(((((......)))))--.)).))..))).).)))))...))))))).. ( -19.40) >DroSec_CAF1 49250 76 - 1 AAAGCAAAAUGAACACACA--------------------------CA---UAGGUAGAAAGGCAAAUACGCCUU--GACUCUCGGUCAGAUAUUCUGAUUUGUUUGA .((((((............--------------------------..---..(((...(((((......)))))--.)))....(((((.....))))))))))).. ( -16.00) >DroSim_CAF1 48798 76 - 1 AAAACAAAAUGAACACACA--------------------------CA---UAGGUAGAAAGGCAAAUACGCCUU--GACUCUCGGUCAGAUAUUCUGAUUUGUUUGA .((((((............--------------------------..---..(((...(((((......)))))--.)))....(((((.....))))))))))).. ( -16.00) >DroEre_CAF1 45395 91 - 1 AAAGCAAAAUGAACAC----------ACACAUCCCA----AUGGACAUUGUAGGUAGAAAGGCAAAUACGCCUU--GACUCUCGGCCAGAUAUUCUGAUUUGUUUGA .(((((((..(((...----------(((..(((..----..)))...))).(((((((((((......)))))--...)))..))).....)))...))))))).. ( -17.10) >DroYak_CAF1 47157 100 - 1 AAAGCAAAAUGAACACACA-GCACACACACAUCCCA---AAUGGACA---UAGGUAGAAAGGCAAAUACGCCUUGUGACUCUCGGUCAGAUAUUCUGAUUUGUUUGA .((((((........(((.-....((.....(((..---...)))..---...))...(((((......)))))))).......(((((.....))))))))))).. ( -18.70) >DroAna_CAF1 48086 100 - 1 AACAUAAAACGGACAUUCA--CAUACACACAUAUGUAUGUAUGCAUG---UAGGUGAAGAGGCAAACACACCUU--GACUCUUGGUCAGAUAUUCUGAUUUGUUUGA .........((((((((((--(.((((..(((((....)))))..))---)).)))))(((.(((.......))--).)))..((((((.....)))))))))))). ( -24.40) >consensus AAAGCAAAAUGAACACACA_______ACACAUCCCA____AUGGACA___UAGGUAGAAAGGCAAAUACGCCUU__GACUCUCGGUCAGAUAUUCUGAUUUGUUUGA .........((((((.....................................(((...(((((......)))))...)))...((((((.....)))))))))))). (-14.49 = -14.27 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:55 2006