| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,993,061 – 11,993,169 |
| Length | 108 |
| Max. P | 0.506293 |

| Location | 11,993,061 – 11,993,169 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.94 |
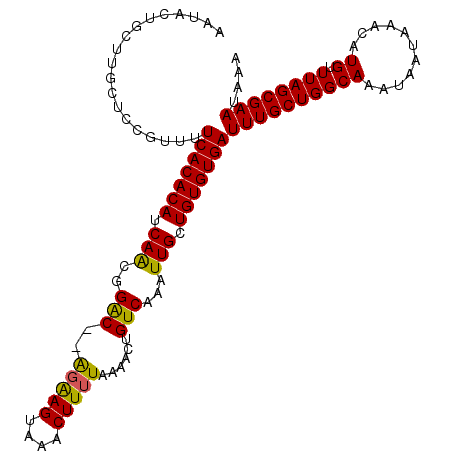
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -19.80 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
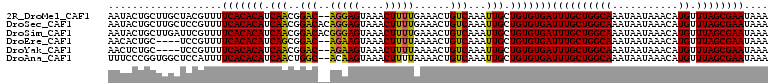
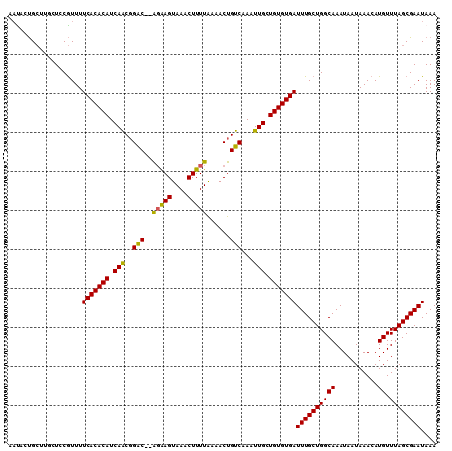
>2R_DroMel_CAF1 11993061 108 + 20766785 AAUACUGCUUGCUACGUUUUCACACAUCAACGGAC--AGGAGUAAACUUUUGAAACUGUCAAAUUGCUGUGUGAUUUGCUGGCAAAUAAUAAACAUGUUUAGCGAAUAAA .....(((((((((.((..(((((((.(((..(((--((...(((....)))...)))))...))).)))))))...))))))))....((((....)))))))...... ( -28.00) >DroSec_CAF1 48317 110 + 1 AAUACUGCUUGCUCCGUUUUCACACAUCAACGGACACAGGAGUAAACUUUUGAAACUGUCAAAUUGCUGUGUGAUUUGCUGGCAAAUAAUAAACAUGUUUAGCGAAUAAA .....((((((((..((..(((((((.(((..((((((((((....))))))....))))...))).)))))))...)).)))))....((((....)))))))...... ( -26.30) >DroSim_CAF1 47837 110 + 1 AAUACUGCUUGAUUCGUUUUCACACAUCAACGGACACGGGAGUAAACUUUUGAAACUGUCAAAUUGCUGUGUGAUUUGCUGGCAAAUAAUAAACAUGUUUAGCGAAUAAA .....((((.(((..(((((((((((.(((..((((((((((....))))))....))))...))).)))))))((((....))))....))))..))).))))...... ( -25.20) >DroEre_CAF1 44435 104 + 1 AACACUGC----UCCGUUUUCACACAUCAGCGGAC--AGAAGUAAACUUUUAAAACUGUCAAAUUGCUGUGUGAUUUGCUGGCAAAUAAUAAACAUGUUUAGCGAAUAAA ........----.......(((((((.(((..(((--((...(((....)))...)))))...))).)))))))((((((((((...........)).)))))))).... ( -22.70) >DroYak_CAF1 46190 104 + 1 AACUCUGC----UCCGUUUUCACACAUCAACGGAC--AGAAGUAAACUUUUAAAACUGUCAAAUUGCUGUGUGAUUUGCUGGCAAAUAAUAAACAUGUUUAGCGAAUAAA ........----.......(((((((.(((..(((--((...(((....)))...)))))...))).)))))))((((((((((...........)).)))))))).... ( -23.00) >DroAna_CAF1 47233 108 + 1 UUUCCCGGUGGCUCCAUUUUCACACAUCAACUGGC--ACAAGUAAACUUUUAAAACUGUCAAAUUGCUGUGUGAUUUGCUGGCAAAUAAUAAACAUGUUUAGCGAAUAAA ......((.....))....(((((((.(((.((((--(....(((....)))....)))))..))).)))))))((((((((((...........)).)))))))).... ( -20.70) >consensus AAUACUGCUUGCUCCGUUUUCACACAUCAACGGAC__AGAAGUAAACUUUUAAAACUGUCAAAUUGCUGUGUGAUUUGCUGGCAAAUAAUAAACAUGUUUAGCGAAUAAA ...................(((((((.(((..(((..(((((....)))))......)))...))).)))))))((((((((((...........)).)))))))).... (-19.80 = -19.30 + -0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:53 2006