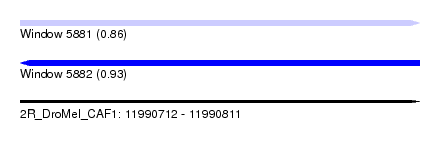
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,990,712 – 11,990,811 |
| Length | 99 |
| Max. P | 0.929941 |

| Location | 11,990,712 – 11,990,811 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.23 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
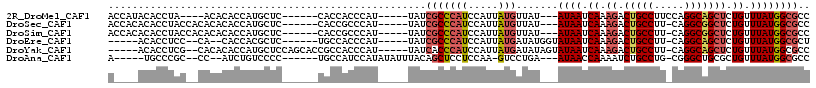
>2R_DroMel_CAF1 11990712 99 + 20766785 GGCGCCAUAAACAGAGCUGCCUGGAAGGCAGUCUUUGAUUAU---AUAACAUAAUGGAUGGGCGAUA-----AUGGGUGGUG------GAGCAUGGUGUGU----UAGGUGUAUGGU .(((((((((.((((((((((.....))))).))))).))))---.(((((((....(((..(.((.-----.......)).------)..)))..)))))----)))))))..... ( -30.00) >DroSec_CAF1 41784 102 + 1 GGCGCCAUAAACAGAGCCGCCUG-AAGGCAGUCUUUGAUUAU---AUAACAUAAUGGAUGGGCGAUA-----AUGGGCGGUG------GAGCAUGGUGUGUGUGGUAGGUGUGUGGU ...((((((.((...(((((((.-......((((((.(((((---.....))))).)).))))....-----..))))))).------..((((.....)))).....)).)))))) ( -27.62) >DroSim_CAF1 45435 102 + 1 GGCGCCAUAAACAGAGCCGCCUG-AAGGCAGUCUUUGAUUAU---AUAACAUAAUGGAUGGGCGAUA-----AUGGGCGGUG------GAGCAUGGUGUGUGUGGUAGGUGUGUGGU ...((((((.((...(((((((.-......((((((.(((((---.....))))).)).))))....-----..))))))).------..((((.....)))).....)).)))))) ( -27.62) >DroEre_CAF1 42139 96 + 1 AGCGCCAUAAACAGAGCUGCCUG-AAGGCAGUCUUUGAUUAUACCAUAUCAUAAUGGAUGGGCGAUA-----AUGGGUGGCA------GAGCGUGGUG--UG--GGAGGUGU----- .(((((((((.((((((((((..-..))))).))))).)))).(((((((((....(....((.((.-----....)).)).------...)))))))--))--)..)))))----- ( -34.80) >DroYak_CAF1 42681 104 + 1 GGCGCCAUAAACAGAGCUGCCUG-AAGGCAGUCUUUGAUUAUACUAUAUCAUAAUGGAUGGGUGAUA-----AUGGGUGGCGGUGCUGGAGCAUGGUGUGUG--CGAGGUGU----- ..((((((...((((((((((..-..))))).)))))......(((((((((.........))))).-----))))))))))(..((...((((.....)))--)..))..)----- ( -35.80) >DroAna_CAF1 45259 97 + 1 GGCGCCAUAAACAGCGCAGCCCG-CAGGCAGAUUUUGGUUAU---UCAGGAC-UUGGAGGAGCUGUAAAUAUAUGGAUGGCA------GGGGACAGAU--GG--GCGGGCA-----U .((((........)))).(((((-(............(((((---(((..(.-(((.((...)).))).)...)))))))).------(....)....--..--)))))).-----. ( -30.00) >consensus GGCGCCAUAAACAGAGCUGCCUG_AAGGCAGUCUUUGAUUAU___AUAACAUAAUGGAUGGGCGAUA_____AUGGGUGGCG______GAGCAUGGUGUGUG__GUAGGUGU____U .(((((.........((((((((.......((((((.(((((........))))).)).))))..........)))))))).........((.....))........)))))..... (-16.31 = -16.23 + -0.08)

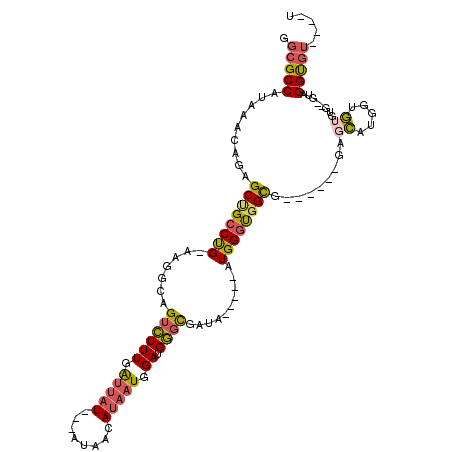
| Location | 11,990,712 – 11,990,811 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.47 |
| Mean single sequence MFE | -18.65 |
| Consensus MFE | -10.89 |
| Energy contribution | -11.62 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11990712 99 - 20766785 ACCAUACACCUA----ACACACCAUGCUC------CACCACCCAU-----UAUCGCCCAUCCAUUAUGUUAU---AUAAUCAAAGACUGCCUUCCAGGCAGCUCUGUUUAUGGCGCC ............----.............------..........-----...(((((((.....)))....---((((.((.((.(((((.....))))))).)).)))))))).. ( -16.10) >DroSec_CAF1 41784 102 - 1 ACCACACACCUACCACACACACCAUGCUC------CACCGCCCAU-----UAUCGCCCAUCCAUUAUGUUAU---AUAAUCAAAGACUGCCUU-CAGGCGGCUCUGUUUAUGGCGCC .........................((..------....))....-----...(((((((.....)))....---((((.((.((.(((((..-..))))))).)).)))))))).. ( -15.60) >DroSim_CAF1 45435 102 - 1 ACCACACACCUACCACACACACCAUGCUC------CACCGCCCAU-----UAUCGCCCAUCCAUUAUGUUAU---AUAAUCAAAGACUGCCUU-CAGGCGGCUCUGUUUAUGGCGCC .........................((..------....))....-----...(((((((.....)))....---((((.((.((.(((((..-..))))))).)).)))))))).. ( -15.60) >DroEre_CAF1 42139 96 - 1 -----ACACCUCC--CA--CACCACGCUC------UGCCACCCAU-----UAUCGCCCAUCCAUUAUGAUAUGGUAUAAUCAAAGACUGCCUU-CAGGCAGCUCUGUUUAUGGCGCU -----........--..--..........------.(((..((((-----.((((...........)))))))).((((.((.((.(((((..-..))))))).)).)))))))... ( -20.60) >DroYak_CAF1 42681 104 - 1 -----ACACCUCG--CACACACCAUGCUCCAGCACCGCCACCCAU-----UAUCACCCAUCCAUUAUGAUAUAGUAUAAUCAAAGACUGCCUU-CAGGCAGCUCUGUUUAUGGCGCC -----.......(--(........(((....)))..(((......-----(((((...........)))))....((((.((.((.(((((..-..))))))).)).))))))))). ( -22.10) >DroAna_CAF1 45259 97 - 1 A-----UGCCCGC--CC--AUCUGUCCCC------UGCCAUCCAUAUAUUUACAGCUCCUCCAA-GUCCUGA---AUAACCAAAAUCUGCCUG-CGGGCUGCGCUGUUUAUGGCGCC .-----.((((((--..--....((....------.))........((((((..(((......)-))..)))---)))..............)-))))).(((((......))))). ( -21.90) >consensus A____ACACCUAC__CACACACCAUGCUC______CACCACCCAU_____UAUCGCCCAUCCAUUAUGUUAU___AUAAUCAAAGACUGCCUU_CAGGCAGCUCUGUUUAUGGCGCC .....................................................(((((((.....))).......((((.((.((.(((((.....))))))).)).)))))))).. (-10.89 = -11.62 + 0.72)
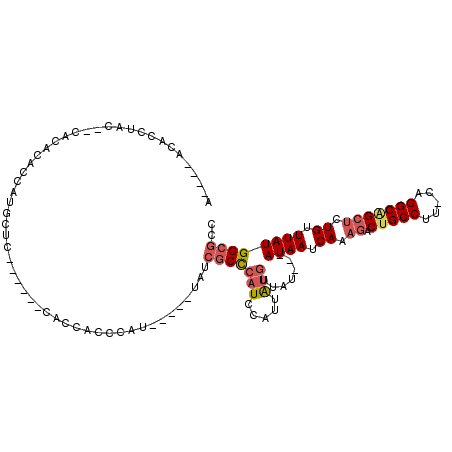
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:52 2006