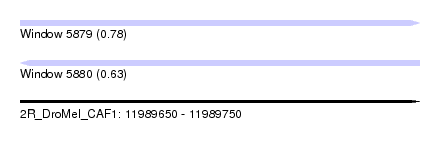
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,989,650 – 11,989,750 |
| Length | 100 |
| Max. P | 0.775898 |

| Location | 11,989,650 – 11,989,750 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.61 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -14.90 |
| Energy contribution | -16.93 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
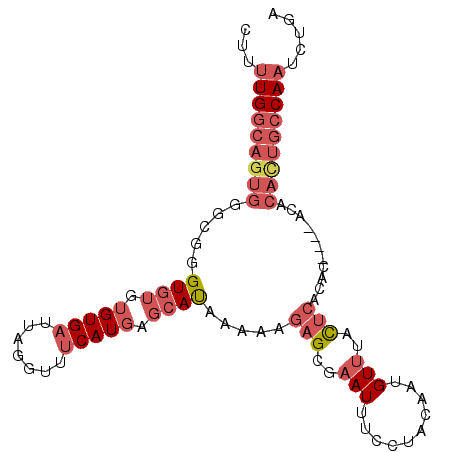
>2R_DroMel_CAF1 11989650 100 + 20766785 UCAGAUUGGCAGUGUGUCUGUGUGUGAGUAAACAUUGUAGGAAAUUCGCUCUUUUUAUGCUCAUGAAACCUAAUCACACACACCCGCCCACUGCCAAAAG .....(((((((((.((.(((((((((..........((((..((..((.........))..))....)))).)))))))))...)).)))))))))... ( -31.40) >DroSec_CAF1 40725 98 + 1 UCAGAUUGGCAGUGUGU--GUGUGUGAGUAAACAUUGUAGGAAAUUCGCUCUUUUUAUGCUCAUGAAACCUAAUCACACACACCCGCCCACUGCCAAAAG .....(((((((((.((--((((((((..........((((..((..((.........))..))....)))).)))))))))).....)))))))))... ( -31.70) >DroSim_CAF1 44368 98 + 1 UCAGAUUGGCAGUGUGU--GUGUGUGAGUAAACAUUGUAUGAAAUUCGCUCUUUUUAUGCUCAUGAAACCUAAUCACACACACCCGCCCACUGCCAAAAG .....(((((((((.((--((((((((.....(((.((((((((........))))))))..)))........)))))))))).....)))))))))... ( -35.42) >DroEre_CAF1 41069 94 + 1 CCAGAUUGGCAAUGU------GUGUGAGUAAACAUUGUAGGAAAUUCCCUCUUUUUAUGCUCAUGAAACCUAAUCACACACACCCGCCCACUGCCAACAG .(((...(((..(((------((((((...........(((......)))..((((((....)))))).....)))))))))...)))..)))....... ( -22.00) >DroYak_CAF1 41571 94 + 1 UCAGAUUGGCAGUGU------GUGUGAGUAAACAUUGUAGGAAAUUCCCUCUUUUUAUGCUCAUGAAACCUAAUCACACACACCCGCCCACUGCCAACAG .(((...(((.((((------((((((...........(((......)))..((((((....)))))).....))))))))))..)))..)))....... ( -25.30) >DroAna_CAF1 44206 76 + 1 ------UGGAAAUGUAU------UU--AUAUACAUUGUAGAAAAUUCCCACAUUUUGUGCUGAUGGUACCUAAUCACACCUACG----------CAAAAU ------....(((((((------..--..)))))))(((((((((......)))))(((.((((........))))))))))).----------...... ( -12.50) >consensus UCAGAUUGGCAGUGUGU____GUGUGAGUAAACAUUGUAGGAAAUUCCCUCUUUUUAUGCUCAUGAAACCUAAUCACACACACCCGCCCACUGCCAAAAG .....(((((((((.......((((((.....(((.(((.((((........)))).)))..)))........)))))).........)))))))))... (-14.90 = -16.93 + 2.03)



| Location | 11,989,650 – 11,989,750 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.61 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -16.82 |
| Energy contribution | -18.77 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11989650 100 - 20766785 CUUUUGGCAGUGGGCGGGUGUGUGUGAUUAGGUUUCAUGAGCAUAAAAAGAGCGAAUUUCCUACAAUGUUUACUCACACACAGACACACUGCCAAUCUGA ...(((((((((...(..(((((((((.((((....((..((.........))..))..))))....(....))))))))))..).)))))))))..... ( -31.50) >DroSec_CAF1 40725 98 - 1 CUUUUGGCAGUGGGCGGGUGUGUGUGAUUAGGUUUCAUGAGCAUAAAAAGAGCGAAUUUCCUACAAUGUUUACUCACACAC--ACACACUGCCAAUCUGA ...(((((((((.....((((((((((.((((....((..((.........))..))..))))....(....)))))))))--)).)))))))))..... ( -32.90) >DroSim_CAF1 44368 98 - 1 CUUUUGGCAGUGGGCGGGUGUGUGUGAUUAGGUUUCAUGAGCAUAAAAAGAGCGAAUUUCAUACAAUGUUUACUCACACAC--ACACACUGCCAAUCUGA ...(((((((((.....((((((((((...(....).(((((((.....(((.....))).....))))))).))))))))--)).)))))))))..... ( -32.70) >DroEre_CAF1 41069 94 - 1 CUGUUGGCAGUGGGCGGGUGUGUGUGAUUAGGUUUCAUGAGCAUAAAAAGAGGGAAUUUCCUACAAUGUUUACUCACAC------ACAUUGCCAAUCUGG .......(((..(((((.(((((((((...(....).(((((((......((((....))))...))))))).))))))------))))))))...))). ( -28.20) >DroYak_CAF1 41571 94 - 1 CUGUUGGCAGUGGGCGGGUGUGUGUGAUUAGGUUUCAUGAGCAUAAAAAGAGGGAAUUUCCUACAAUGUUUACUCACAC------ACACUGCCAAUCUGA .......(((..(((((.(((((((((...(....).(((((((......((((....))))...))))))).))))))------))))))))...))). ( -30.40) >DroAna_CAF1 44206 76 - 1 AUUUUG----------CGUAGGUGUGAUUAGGUACCAUCAGCACAAAAUGUGGGAAUUUUCUACAAUGUAUAU--AA------AUACAUUUCCA------ ((((((----------.((.((((.(........))))).)).))))))((((((...))))))(((((((..--..------)))))))....------ ( -15.00) >consensus CUUUUGGCAGUGGGCGGGUGUGUGUGAUUAGGUUUCAUGAGCAUAAAAAGAGCGAAUUUCCUACAAUGUUUACUCACAC____ACACACUGCCAAUCUGA ...(((((((((.....((((.(((((.......))))).)))).....(((..(((..........)))..)))...........)))))))))..... (-16.82 = -18.77 + 1.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:50 2006