
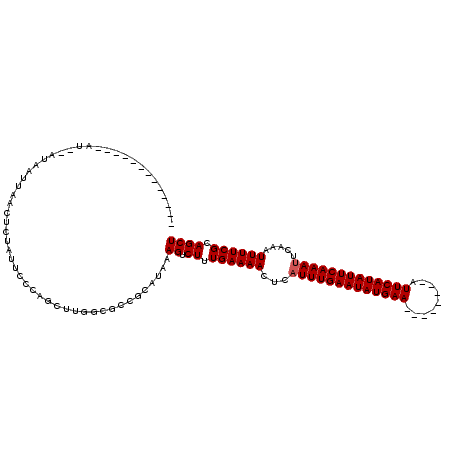
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,985,871 – 11,985,966 |
| Length | 95 |
| Max. P | 0.961431 |

| Location | 11,985,871 – 11,985,966 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.86 |
| Mean single sequence MFE | -17.35 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
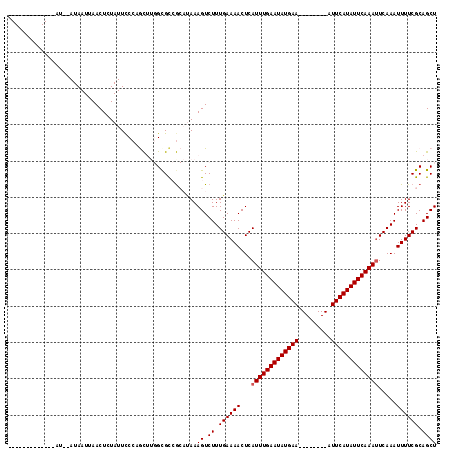
>2R_DroMel_CAF1 11985871 95 + 20766785 -------------AU--AUAAUUAACUAUAUUCCCAGCUUGGCGCCGCAUAAAGUCUUUGAAAACUCAUUUGAAUAUGAA--------AUUCAUAUUCAAAUUCAAAUUUUCGCAGCU -------------..--..................((((.(((..........)))...(((((...(((((((((((..--------...))))))))))).....)))))..)))) ( -16.10) >DroVir_CAF1 40117 114 + 1 UGUCUAUAUACAUAU--AUAAUUAACUCUAUUUCCCACAA-GCGCGG-CCAAAGUCUUUGAAAACUCAUUUGAAUAUGAAAUUCUGAAAUUCAUAUUCAAAUUCAAAUUUUCGCAGCU ....(((((....))--)))...................(-((((((-(....)))...(((((...(((((((((((((.........))))))))))))).....))))))).))) ( -18.90) >DroGri_CAF1 37676 115 + 1 UGUCUAUAUACAUAUACAUAAUUAACUCUAUUCCC-ACAA-GCGUGG-CUAAAGUCUUUGAAAACUCAUUUGAAUAUGAAAUUCUGAAAUUCAUAUUCAAAUUCAAAUUUUCGCAGCU .................................((-((..-..))))-....((.((.((((((...(((((((((((((.........))))))))))))).....)))))).)))) ( -19.00) >DroSec_CAF1 37080 95 + 1 -------------AU--AUAAUUAACUGUAUUCCCAGCUUGGCGCCGCAUAAAGUCUUUGAAAACUCAUUUGAAUAUGAA--------AUUCAUAUUCAAAAUCAAAUUUUCGCAGCU -------------..--........((((.......((((.((...))...))))....(((((....((((((((((..--------...))))))))))......))))))))).. ( -16.60) >DroYak_CAF1 37829 95 + 1 -------------AU--AUAAUUAACUGUAUUCCCAGCUUGGUGCCGCAUAAAGUCUUUGAAAACUCAUUUGAAUAUGAA--------AUUCAUAUUCAAAUUCAAAUUUUCGCAGCU -------------..--........((((.......((((.(((...))).))))....(((((...(((((((((((..--------...))))))))))).....))))))))).. ( -17.50) >DroAna_CAF1 40636 94 + 1 -------------AU--AUAAUUAACUCUAUUCCC-GUUUGGCGCCACACAAAGUCUUUGAAAACUCAUUUGAAUAUGAA--------AUUCAUAUUCAAAUUCAAAUUUUCGCAGCU -------------..--..................-((.((....)).))..((.((.((((((...(((((((((((..--------...))))))))))).....)))))).)))) ( -16.00) >consensus _____________AU__AUAAUUAACUCUAUUCCCAGCUUGGCGCCGCAUAAAGUCUUUGAAAACUCAUUUGAAUAUGAA________AUUCAUAUUCAAAUUCAAAUUUUCGCAGCU ....................................................((.((.((((((...(((((((((((((.........))))))))))))).....)))))).)))) (-14.22 = -14.38 + 0.17)

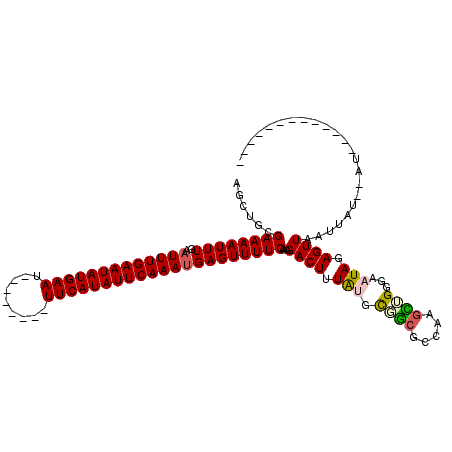
| Location | 11,985,871 – 11,985,966 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.86 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -21.80 |
| Energy contribution | -21.92 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
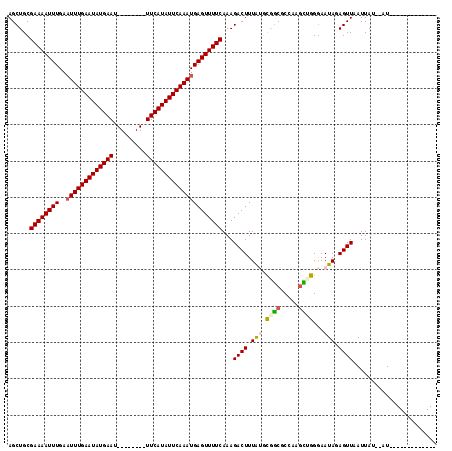
>2R_DroMel_CAF1 11985871 95 - 20766785 AGCUGCGAAAAUUUGAAUUUGAAUAUGAAU--------UUCAUAUUCAAAUGAGUUUUCAAAGACUUUAUGCGGCGCCAAGCUGGGAAUAUAGUUAAUUAU--AU------------- ......((((((((..(((((((((((...--------..)))))))))))))))))))...((((.(((.((((.....))))...))).))))......--..------------- ( -25.20) >DroVir_CAF1 40117 114 - 1 AGCUGCGAAAAUUUGAAUUUGAAUAUGAAUUUCAGAAUUUCAUAUUCAAAUGAGUUUUCAAAGACUUUGG-CCGCGC-UUGUGGGAAAUAGAGUUAAUUAU--AUAUGUAUAUAGACA ......((((((((..(((((((((((((.........)))))))))))))))))))))...(((((((.-((((..-..))))....)))))))..((((--((....))))))... ( -28.20) >DroGri_CAF1 37676 115 - 1 AGCUGCGAAAAUUUGAAUUUGAAUAUGAAUUUCAGAAUUUCAUAUUCAAAUGAGUUUUCAAAGACUUUAG-CCACGC-UUGU-GGGAAUAGAGUUAAUUAUGUAUAUGUAUAUAGACA ......((((((((..(((((((((((((.........)))))))))))))))))))))...(((((((.-((((..-..))-))...)))))))..(((((((....)))))))... ( -30.90) >DroSec_CAF1 37080 95 - 1 AGCUGCGAAAAUUUGAUUUUGAAUAUGAAU--------UUCAUAUUCAAAUGAGUUUUCAAAGACUUUAUGCGGCGCCAAGCUGGGAAUACAGUUAAUUAU--AU------------- .(((((((((((((...((((((((((...--------..)))))))))).))))))))((.....))..)))))....(((((......)))))......--..------------- ( -26.50) >DroYak_CAF1 37829 95 - 1 AGCUGCGAAAAUUUGAAUUUGAAUAUGAAU--------UUCAUAUUCAAAUGAGUUUUCAAAGACUUUAUGCGGCACCAAGCUGGGAAUACAGUUAAUUAU--AU------------- .((((((.........(((((((((((...--------..)))))))))))((((((....))))))..))))))....(((((......)))))......--..------------- ( -27.30) >DroAna_CAF1 40636 94 - 1 AGCUGCGAAAAUUUGAAUUUGAAUAUGAAU--------UUCAUAUUCAAAUGAGUUUUCAAAGACUUUGUGUGGCGCCAAAC-GGGAAUAGAGUUAAUUAU--AU------------- ......((((((((..(((((((((((...--------..)))))))))))))))))))...((((((((....(.((....-))).))))))))......--..------------- ( -24.30) >consensus AGCUGCGAAAAUUUGAAUUUGAAUAUGAAU________UUCAUAUUCAAAUGAGUUUUCAAAGACUUUAUGCGGCGCCAAGCUGGGAAUAGAGUUAAUUAU__AU_____________ ......((((((((..(((((((((((((.........)))))))))))))))))))))...((((.(((.((((.....))))...))).))))....................... (-21.80 = -21.92 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:44 2006