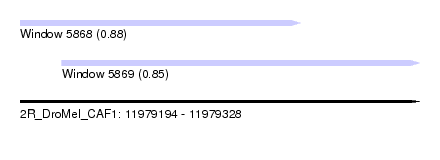
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,979,194 – 11,979,328 |
| Length | 134 |
| Max. P | 0.879725 |

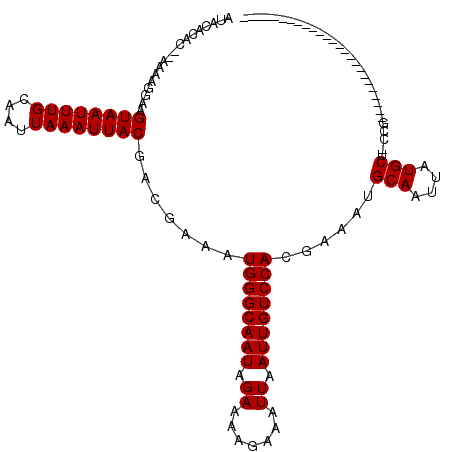
| Location | 11,979,194 – 11,979,288 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11979194 94 + 20766785 AAUUAC-GUC-------------------------UACAAAUACACAUAAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGGAAUUAAUUGUCCACGAAAU .....(-(((-------------------------..........(......).....((((((((....))))))))))))...((((((((.((.......)).))))))))...... ( -16.00) >DroVir_CAF1 35801 112 + 1 AAUUAC-GGGCGAA-----GGCAGCGCAGCAAACGGACACAAACACAC--AAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAAUUAAUUGUCCACGAAAU .....(-(.((...-----.))..)).......((.............--........((((((((....)))))))).......((((((((.((.......)).)))))))))).... ( -18.20) >DroGri_CAF1 33632 113 + 1 AAUUACUGGGCAAA-----GUCAGUGUGGCAAAUGGACACACAAACAC--AAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAAUUAAUUGUCCACGAAAU ..............-----(((.(((((.........)))))......--........((((((((....)))))))))))....((((((((.((.......)).))))))))...... ( -23.00) >DroEre_CAF1 30669 94 + 1 AAUUAC-GUC-------------------------UACAAAUACACAUAAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAGGGAAUUAAUUGUCCACGAAAU .....(-(((-------------------------..........(......).....((((((((....))))))))))))...((((((((.((.......)).))))))))...... ( -16.00) >DroAna_CAF1 31584 94 + 1 AAUUAC-GUC-------------------------UACAAAUACACAUAAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAGGGAAUUAAUUGUCCACGAAAU .....(-(((-------------------------..........(......).....((((((((....))))))))))))...((((((((.((.......)).))))))))...... ( -16.00) >DroMoj_CAF1 40455 115 + 1 AAUUGC-GAACGAGCAGCGAGCAGCGCAGCAAACGGACACAAACA--C--AAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAAUUAAUUGUCCACGAAAU ..(((.-(..((.((.(((.....))).))...))..).)))...--.--........((((((((....)))))))).......((((((((.((.......)).))))))))...... ( -20.60) >consensus AAUUAC_GUC_________________________GACAAAUACACAC__AAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAAUUAAUUGUCCACGAAAU ..........................................................((((((((....)))))))).......((((((((.((.......)).))))))))...... (-12.60 = -12.60 + -0.00)

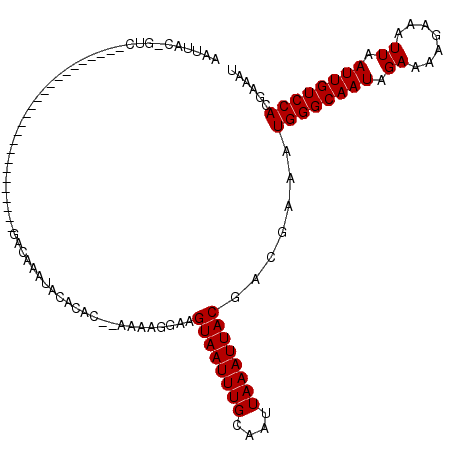
| Location | 11,979,208 – 11,979,328 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11979208 120 + 20766785 AUACACAUAAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGGAAUUAAUUGUCCACGAAAUGCAAUUAUGCUUUCCCCUCUCUUUUUCCGCCUUUUCUUUU ........((((((((((((((((((....))))))))((.((..((((((((.((.......)).)))))))).((((.(((....)))))))...))))...)))).))))))..... ( -26.70) >DroVir_CAF1 35835 92 + 1 AAACACAC--AAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAAUUAAUUGUCCACGAAAUGCAAUUAUGCU-CCG------------------------- ........--....(((.((((((((....)))))))).......((((((((.((.......)).))))))))......(((....))))-)).------------------------- ( -17.00) >DroGri_CAF1 33667 92 + 1 ACAAACAC--AAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAAUUAAUUGUCCACGAAAUGCAAUUAUGCU-CCG------------------------- ........--....(((.((((((((....)))))))).......((((((((.((.......)).))))))))......(((....))))-)).------------------------- ( -17.00) >DroSec_CAF1 30398 117 + 1 AUACACAUAAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGGAAUUAAUUGUCCACGAAAUGCAAUUAUGCU-UCCCGUCGCUUAUUCCGCAUUUC--UUU ............(.((((((((((((....))))))))((((...((((((((.((.......)).))))))))......(((....))).-...)))).....)))).).....--... ( -26.00) >DroAna_CAF1 31598 104 + 1 AUACACAUAAAAGGGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAGGGAAUUAAUUGUCCACGAAAUGCAAUUAUGCU-CAGC-UUUCUUACU-------------- ..........(((..(..((((((((....))))))))...((..((((((((.((.......)).))))))))......(((....))))-)...-)..)))...-------------- ( -17.00) >DroMoj_CAF1 40494 90 + 1 AAACA--C--AAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAAUUAAUUGUCCACGAAAUGCAAUUAUGCU-CCG------------------------- .....--.--....(((.((((((((....)))))))).......((((((((.((.......)).))))))))......(((....))))-)).------------------------- ( -17.00) >consensus AUACACAC__AAAAGGAAGUAAUUUGCAAUUAAAUUACGACGAAAUGGGCAAUAGAAAAGAAAUUAAUUGUCCACGAAAUGCAAUUAUGCU_CCG_________________________ ..................((((((((....)))))))).......((((((((.((.......)).))))))))......(((....))).............................. (-14.00 = -14.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:40 2006