| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,978,326 – 11,978,428 |
| Length | 102 |
| Max. P | 0.835426 |

| Location | 11,978,326 – 11,978,428 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
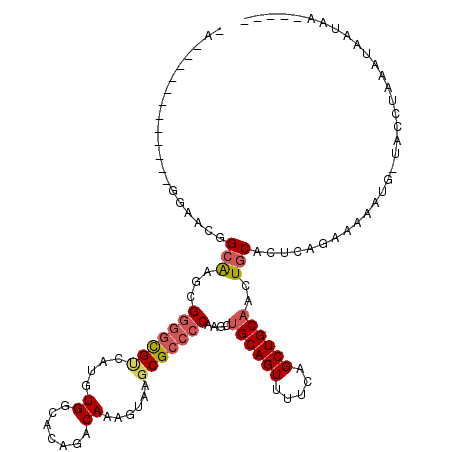
>2R_DroMel_CAF1 11978326 102 - 20766785 GG------------GGAAUGGCAAGCGGGGCGUCAUGUGGCACAGACAAAGUAAGCGCCCCAAGCUGCAGUUUUCAGCUGCAUCUGCACUCAGAAAAAUG-UACGGAAAUAAUAA----- ((------------(.....(((((((((((((....((.......))......)))))))..)))(((((.....)))))...))).))).........-..............----- ( -27.80) >DroSec_CAF1 29526 101 - 1 -G------------GCAACGGCAAGCGGGCCGUCAUGUGGCACAGACAAAGUAAGCGCCCCAAACUGCAGUUUUCAGCUGCAUCUGCACUCAGAAAAAUG-UACGGAAGUAAUAA----- -(------------((...(((......)))(((.((.....))))).........)))......((((((.....))))))((((....))))......-(((....)))....----- ( -26.00) >DroSim_CAF1 33069 95 - 1 -------------------GGCAGGCGGGGCGCCAUGUGGCACAGACAAAGUAAGCGCCCCAAGCUGCAGUUUUCAGCUGCAUCUGCACUCAGAAAAAUU-UACGAAAGUAAUAA----- -------------------.(((((.(((((((....((.......))......)))))))....((((((.....)))))))))))............(-(((....))))...----- ( -34.80) >DroEre_CAF1 29837 101 - 1 -A------------GGAACGGCAAACGGGGCGUCAUGUGGCACAGACAAAGUAAGCGCCCCAAGCUGCAGUUUCCAGCUGCAACUGCACUCAGAAAAAGG-UACCUCCAUAACAA----- -.------------(((...((....(((((((....((.......))......)))))))..((((((((.....))))))...))............)-)...))).......----- ( -28.70) >DroYak_CAF1 30220 101 - 1 -A------------GGAGCUGGAAGCGGAGCGUCAUGUGGCACAGACAAAGUAAGCGCCCCAAGCUGCAGUUUCCAGCUGCAACUGCACUAAGAAAAAGG-UACCUCCAUAAGAG----- -.------------(((((((((((((.(((....((.(((.(..((...))..).))).)).))).).)))))))))(((....)))............-....))).......----- ( -30.30) >DroAna_CAF1 30635 119 - 1 GACACGCGGACUCUGCAGGGGCGAGUG-GGUGGCAUGUGGCACAGACAAAGUAAGCGCCCCAAGCUGCAGUUUGCAGCUGCCAGUGCACUGAACAAAACAAGAACUGCAAAGCAAAUACA .....((((..(((((.((((((..((-.(((.(....).)))...)).......)))))).((((((.....))))))))(((....))).........))).))))............ ( -37.70) >consensus _A____________GGAACGGCAAGCGGGGCGUCAUGUGGCACAGACAAAGUAAGCGCCCCAAGCUGCAGUUUUCAGCUGCAACUGCACUCAGAAAAAUG_UACCUAAAUAAUAA_____ ....................(((...(((((((....((.......))......)))))))....((((((.....))))))..)))................................. (-19.50 = -20.12 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:37 2006