| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,977,648 – 11,977,746 |
| Length | 98 |
| Max. P | 0.979449 |

| Location | 11,977,648 – 11,977,746 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -11.79 |
| Energy contribution | -11.70 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11977648 98 + 20766785 U-UUCGGUCGGCA-AGUGCAGGUUGCAAAAUAAUGAAUGGGGCCUGCAUUUAUUAAUGUAUUUGGAAAGCGU-UCGCAAAUUAAUUGCCAUUUAAUUUGUU .-.......((((-((((((((((.((..........)).)))))))))))........((((((((....)-)).))))).....)))............ ( -24.40) >DroVir_CAF1 34001 86 + 1 U-UUUGG-CUAUU-AAUGCAGGU------GUAAUGAAUG-UGCCUGCAUUUAUUAAUGCUUU----CAACUU-UUGCAAAUUAAUUGGCAUUUAAUUUAUA .-....(-(((..-((((((((.------.((.....))-..))))))))......(((...----......-..))).......))))............ ( -19.40) >DroGri_CAF1 32055 86 + 1 UUUUCGA-CUAUA-AAUGCAGGU------GUAAUGGAUG-UGCCUGCAUUUAUUAAUGCUUC----AAACUU--UCCAAAUUAAUUGGCAUUUAAUUUAUU .......-..(((-((((((((.------.((.....))-..))))))))))).((((((..----......--............))))))......... ( -20.25) >DroYak_CAF1 29565 100 + 1 U-UUCGGUCGGCAAAGUGCAGGUUGCAAAAUAAUGAAUGGGGCCUGCAUUUAUUAAUGUAUUUGGAAAGCGUUUCGCAAAUUAAUUGCCAUUUAAUUUGUU .-.......(((((((((((((((.((..........)).)))))))))))........((((((((.....))).)))))....))))............ ( -25.30) >DroAna_CAF1 30016 97 + 1 U-UUUGGUCGGCA-AGUGCAGGUUGCAA-AUAAUGAAUGGGGCCUGCAUUUAUUAAUGUUUUUGGAAAGCGU-UCGCAAAUUAAUUGCCAUUUAAUUUGUU .-.......((((-((((((((((.((.-........)).)))))))))))...(((((((.....))))))-)............)))............ ( -24.40) >DroMoj_CAF1 38117 86 + 1 U-UUUGG-CUAUU-AAUGCAGGU------GUAAUGAAUG-UGCCUGCAUUUAUUAAUGCUGC----CAACUU-UUGCUAAUUAAUUGCCAUUUAAUUUAUA .-..(((-(....-((((((((.------.((.....))-..)))))))).((((((...((----......-..))..)))))).))))........... ( -22.20) >consensus U_UUCGG_CGACA_AAUGCAGGU______AUAAUGAAUG_GGCCUGCAUUUAUUAAUGCUUU____AAACGU_UCGCAAAUUAAUUGCCAUUUAAUUUAUU ..............(((((((((..................)))))))))...........................(((((((.......)))))))... (-11.79 = -11.70 + -0.08)

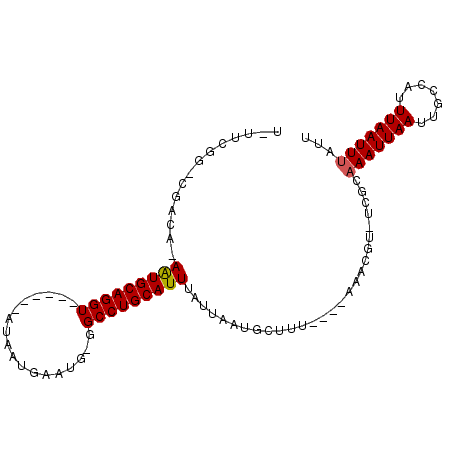
| Location | 11,977,648 – 11,977,746 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -18.16 |
| Consensus MFE | -9.57 |
| Energy contribution | -9.48 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11977648 98 - 20766785 AACAAAUUAAAUGGCAAUUAAUUUGCGA-ACGCUUUCCAAAUACAUUAAUAAAUGCAGGCCCCAUUCAUUAUUUUGCAACCUGCACU-UGCCGACCGAA-A ...........((((((...(((((.((-(....))))))))...........((((((...((..........))...)))))).)-)))))......-. ( -18.20) >DroVir_CAF1 34001 86 - 1 UAUAAAUUAAAUGCCAAUUAAUUUGCAA-AAGUUG----AAAGCAUUAAUAAAUGCAGGCA-CAUUCAUUAC------ACCUGCAUU-AAUAG-CCAAA-A .........(((((...(((((((....-))))))----)..)))))....((((((((..-..........------.))))))))-.....-.....-. ( -16.72) >DroGri_CAF1 32055 86 - 1 AAUAAAUUAAAUGCCAAUUAAUUUGGA--AAGUUU----GAAGCAUUAAUAAAUGCAGGCA-CAUCCAUUAC------ACCUGCAUU-UAUAG-UCGAAAA ......((((((.((((.....)))).--..))))----)).......(((((((((((..-..........------.))))))))-)))..-....... ( -19.82) >DroYak_CAF1 29565 100 - 1 AACAAAUUAAAUGGCAAUUAAUUUGCGAAACGCUUUCCAAAUACAUUAAUAAAUGCAGGCCCCAUUCAUUAUUUUGCAACCUGCACUUUGCCGACCGAA-A ...........((((((...(((((.(((.....))))))))...........((((((...((..........))...))))))..))))))......-. ( -18.40) >DroAna_CAF1 30016 97 - 1 AACAAAUUAAAUGGCAAUUAAUUUGCGA-ACGCUUUCCAAAAACAUUAAUAAAUGCAGGCCCCAUUCAUUAU-UUGCAACCUGCACU-UGCCGACCAAA-A ...........((((((....((((.((-(....)))))))............((((((...((........-.))...)))))).)-)))))......-. ( -17.30) >DroMoj_CAF1 38117 86 - 1 UAUAAAUUAAAUGGCAAUUAAUUAGCAA-AAGUUG----GCAGCAUUAAUAAAUGCAGGCA-CAUUCAUUAC------ACCUGCAUU-AAUAG-CCAAA-A ...........((((.((((((..((..-......----))...)))))).((((((((..-..........------.))))))))-....)-)))..-. ( -18.52) >consensus AACAAAUUAAAUGGCAAUUAAUUUGCAA_AAGCUU____AAAACAUUAAUAAAUGCAGGCA_CAUUCAUUAC______ACCUGCACU_UACAG_CCAAA_A ..((((((((.......))))))))............................((((((....................))))))................ ( -9.57 = -9.48 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:34 2006