
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,973,483 – 11,973,588 |
| Length | 105 |
| Max. P | 0.740467 |

| Location | 11,973,483 – 11,973,588 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -13.34 |
| Energy contribution | -13.37 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11973483 105 - 20766785 GGAUCCAAUCC-GUAGUC-----UCUUCAUCAAAAGUCUGCCAA-AGAGUGGCAACUCUGACAGGCG--GGAUCUGUCAUAAAAUGCACA-AAAAAAAA-----AACAAAAAAACAGAGU ((((...))))-.....(-----(((..........((((((..-(((((....)))))....))))--))...((((((...))).)))-........-----...........)))). ( -19.40) >DroSec_CAF1 24628 96 - 1 GGAUCCAAUCC-GUAGUC-----UCUUCAUCAAAUGUCUGCCAA-AGAGUGGCAACUCUGACAGGCG--GGAUCUGUCAUAAAACGCACA-AAAAAAGA-----AA---------AUAGU ((((((.....-...(..-----....)......((((((....-(((((....)))))..))))))--))))))...............-........-----..---------..... ( -18.80) >DroSim_CAF1 28235 96 - 1 GGAUCCAAUCC-GUAGUC-----UCUUCAUCAAAUGUCUGCCAA-AGAGUGGCAACUCUGACAGGCG--GGAUCUGUCAUAAAACGCACA-AAAAAAGA-----AA---------AGAGU ((((...))))-.....(-----((((..((.....((((((..-(((((....)))))....))))--))...(((........)))..-......))-----.)---------)))). ( -20.00) >DroEre_CAF1 24833 104 - 1 GGAUCCAAUCC-GUAGUC-----UCCUCAUCAAAUGUCUGCAAA-AGAGUGGCAACUCUGACAGCCA--GGAUCUGUCAAAAAACGCACA-ACCAAAAAAAAAAAACAAAAA------AA ((((((.....-((((.(-----............).))))...-(((((....)))))........--))))))...............-.....................------.. ( -17.50) >DroYak_CAF1 25207 109 - 1 GGAUCCAAUCC-GUAGUC-----UCUUCAUCAAAUGUCUGCGAA-AGAGUGGCAACUCUGACAGCCAGAGGAUCUGUCAUAAAACGCACA-AACAAAAAUAAAAAAAAAAAA---CCAAA ((((((....(-((((.(-----............).)))))..-(((((....)))))..........))))))...............-.....................---..... ( -19.30) >DroAna_CAF1 25587 113 - 1 GGAUCCAAAACGGUGGGCGGCGGUCUUCAUCAAAUGUCUGCCAAAAAAGUGGCAACUCUGAACAGGA--GGAUCUGUCAAAAGCCCUGAAAAUGAAAAAAAGGAAAGAAA-----AAAGU ...(((........((((((((((((((.(((...((.(((((......)))))))..)))....))--))).)))))....))))......(....)...)))......-----..... ( -26.90) >consensus GGAUCCAAUCC_GUAGUC_____UCUUCAUCAAAUGUCUGCCAA_AGAGUGGCAACUCUGACAGGCA__GGAUCUGUCAUAAAACGCACA_AAAAAAAA_____AA_AAA_____AGAGU ((((((......((((.....................))))....(((((....)))))..........))))))............................................. (-13.34 = -13.37 + 0.03)

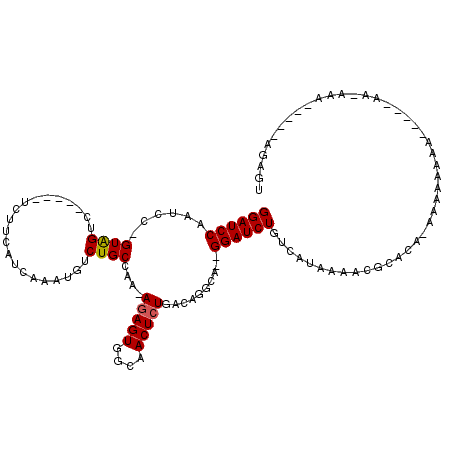
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:32 2006