
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,968,541 – 11,968,650 |
| Length | 109 |
| Max. P | 0.929465 |

| Location | 11,968,541 – 11,968,650 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
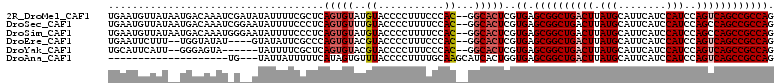
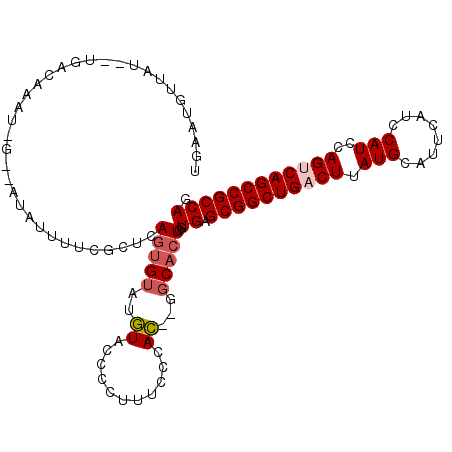
>2R_DroMel_CAF1 11968541 109 + 20766785 UGAAUGUUAUAAUGACAAAUCGAUAUAUUUUCGCUCAGUGUAUGUACCCCUUUCCCAC--GGCACUCGUGAGCGGCUGACUUAUGCAUUCAUCCAUCCAGUCAGCCGCCAG ....((((.....)))).............((((..(((((.(((...........))--)))))).))))((((((((((.(((........)))..))))))))))... ( -27.10) >DroSec_CAF1 19802 109 + 1 UGAAUGUUAUAAUGACAAAUCGGAAUAUUUUCCCUCAGUGUUUGUACCCCUUUUCCAC--GGCACUCGUGAGCGGCUGACUUAUGCAUUCAUCCAUCCAGCCAGCCGCCAG .(((.........((((....((((....)))).....))))..........)))(((--(.....)))).(((((((.((.(((........)))..)).)))))))... ( -23.31) >DroSim_CAF1 23303 109 + 1 UGAAUGUUAUAAUGACAAAUGGGAAUAUUUUCCCUCAGUGUAUGUACCCCUUUUCCAC--GGCACUCGUGAGCGGCUGACUUAUGCAUUCAUCCAUCCAGCCAGCCGCCAG .......((((...(((...(((((....)))))....))).)))).........(((--(.....)))).(((((((.((.(((........)))..)).)))))))... ( -26.70) >DroEre_CAF1 19897 103 + 1 UGAAUUCUUU--UGGUAUAU----GUAUAUUCGCCCAGUGUACGUACCCCUUUCCCAC--GGCACUCGUGAGCGGCUGACUUAUGCAUUCAUCCAUCCAGUCAGCCGCCAG ..........--.((..(((----(((((((.....))))))))))..)).....(((--(.....)))).((((((((((.(((........)))..))))))))))... ( -33.10) >DroYak_CAF1 20148 101 + 1 UGCAUUCAUU--GGGAGUA------UAUUUUCGCUCAGUGUACGUACCCCUUUCCCAC--GGCACUCGUGAGCGGCUGACUUAUGCAUUCAUCCAUCCAGUCAGCCGCCAG (((...((((--(((.(..------......).)))))))...))).........(((--(.....)))).((((((((((.(((........)))..))))))))))... ( -30.70) >DroAna_CAF1 21155 88 + 1 --------------------UG---UAUUAUUUUUCAUAGUGUUUACCCCUUUUGCAAGCAUCACUGGUGAGCGGCUGACUUAUGCAUUCAUCCAUCCAGUCAGCCGCCAG --------------------..---((((((.....)))))).(((((.....((....)).....)))))((((((((((.(((........)))..))))))))))... ( -23.90) >consensus UGAAUGUUAU__UGACAAAU_G__AUAUUUUCGCUCAGUGUAUGUACCCCUUUCCCAC__GGCACUCGUGAGCGGCUGACUUAUGCAUUCAUCCAUCCAGUCAGCCGCCAG ....................................(((((..((...........))...)))))..((.((((((((((.(((........)))..)))))))))))). (-18.88 = -19.43 + 0.56)

| Location | 11,968,541 – 11,968,650 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -25.79 |
| Energy contribution | -25.77 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
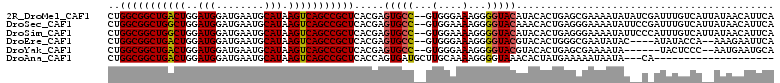
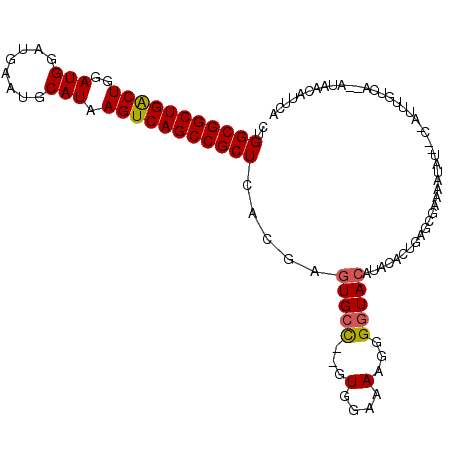
>2R_DroMel_CAF1 11968541 109 - 20766785 CUGGCGGCUGACUGGAUGGAUGAAUGCAUAAGUCAGCCGCUCACGAGUGCC--GUGGGAAAGGGGUACAUACACUGAGCGAAAAUAUAUCGAUUUGUCAUUAUAACAUUCA .((((((((((((..(((........))).))))))))(((((...(((((--.(......).)))))......)))))................))))............ ( -32.90) >DroSec_CAF1 19802 109 - 1 CUGGCGGCUGGCUGGAUGGAUGAAUGCAUAAGUCAGCCGCUCACGAGUGCC--GUGGAAAAGGGGUACAAACACUGAGGGAAAAUAUUCCGAUUUGUCAUUAUAACAUUCA ..(((((((((((..(((........))).))))))))))).(((((((((--.(......).)))))..........((((....))))...)))).............. ( -32.80) >DroSim_CAF1 23303 109 - 1 CUGGCGGCUGGCUGGAUGGAUGAAUGCAUAAGUCAGCCGCUCACGAGUGCC--GUGGAAAAGGGGUACAUACACUGAGGGAAAAUAUUCCCAUUUGUCAUUAUAACAUUCA ..(((((((((((..(((........))).))))))))))).(((((((((--.(......).))))).........(((((....)))))..)))).............. ( -36.30) >DroEre_CAF1 19897 103 - 1 CUGGCGGCUGACUGGAUGGAUGAAUGCAUAAGUCAGCCGCUCACGAGUGCC--GUGGGAAAGGGGUACGUACACUGGGCGAAUAUAC----AUAUACCA--AAAGAAUUCA .(((.((((((((..(((........))).))))))))(((((...(((((--.(......).)))))......)))))........----.....)))--.......... ( -31.80) >DroYak_CAF1 20148 101 - 1 CUGGCGGCUGACUGGAUGGAUGAAUGCAUAAGUCAGCCGCUCACGAGUGCC--GUGGGAAAGGGGUACGUACACUGAGCGAAAAUA------UACUCCC--AAUGAAUGCA ...((((((((((..(((........))).))))))))(((((...(((((--.(......).)))))......))))).......------.......--.......)). ( -32.80) >DroAna_CAF1 21155 88 - 1 CUGGCGGCUGACUGGAUGGAUGAAUGCAUAAGUCAGCCGCUCACCAGUGAUGCUUGCAAAAGGGGUAAACACUAUGAAAAAUAAUA---CA-------------------- ...((((((((((..(((........))).))))))))))(((..((((.(((((.......)))))..)))).))).........---..-------------------- ( -27.10) >consensus CUGGCGGCUGACUGGAUGGAUGAAUGCAUAAGUCAGCCGCUCACGAGUGCC__GUGGAAAAGGGGUACAUACACUGAGCGAAAAUAU__C_AUUUGUCA__AUAACAUUCA ..(((((((((((..(((........))).))))))))))).....(((((...(....)...)))))........................................... (-25.79 = -25.77 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:28 2006