
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,962,479 – 11,962,583 |
| Length | 104 |
| Max. P | 0.795566 |

| Location | 11,962,479 – 11,962,583 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 93.98 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.25 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11962479 104 - 20766785 ------UGCCC-AAUGGCCGCAAGGAAUUUCACACAAUUGCUAGAAAAAUUGCAGAAAAUUGAAAAUGUGCGAGAGCGCAUAAAUUUUUACUGCGUCACUUGUGAAUGCUA ------.(((.-...)))((((((((.((((............))))....((((((((((....((((((....)))))).))))))..)))).)).))))))....... ( -26.70) >DroVir_CAF1 16048 108 - 1 GA--GUCGCCCGAAUGGCCGCAAGGAAUUUCACACAAUUGCUAGAAAAAUUGCAGAAAAUUGAAAAUGUGCGAGAGCGUAUAAAUUUUUACUGCGUCACUUGU-AAUGCUA ..--...((..(((...((....))...))).....(((((.((......(((((((((((....((((((....)))))).))))))..)))))...)).))-))))).. ( -22.50) >DroGri_CAF1 15059 107 - 1 CG--GCAGCCC-AAUGGCCGCAAGGAAUUUCACACAAUUGCUAGAAAAAUUGCAGAAAAUUGAAAAUGUGCGAGAGCAUAUAAAUUUUUACUGCGUCACUUGU-AAUGCUA .(--((((((.-...))).(((((((.((((............))))....((((((((((....((((((....)))))).))))))..)))).)).)))))-..)))). ( -27.20) >DroEre_CAF1 13910 104 - 1 ------UGCCC-AAUGGCCGCAAGGAAUUUCACACAAUUGCUAGAAAAAUUGCAGAAAAUUGAAAAUGUGCGAGAGCGCAUAAAUUUUUACUGCGUCACUUGUGAAUGCUA ------.(((.-...)))((((((((.((((............))))....((((((((((....((((((....)))))).))))))..)))).)).))))))....... ( -26.70) >DroAna_CAF1 14415 110 - 1 UGCCGUUGCCC-AAUGGCCGCAAGGAAUUUCACACAAUUGCUAGAAAAAUUGCAGAAAAUUGAAAAUGUGCGAGAGCGCAUAAAUUUUUACUGCGUCACUUGUGAAUGCUA .((((((....-))))))((((((((.((((............))))....((((((((((....((((((....)))))).))))))..)))).)).))))))....... ( -30.80) >DroMoj_CAF1 16844 107 - 1 UG--CUUGCCC-AAUGGCCGCAAGGAAUUUCACACAAUUGUUAGAAAAAUUGCAGAAAAUUGAAAAUGUGCGAGAGCGUAUAAAUUUUUACUGCGUCACUUGU-AAUGCUA .(--(..(((.-...))).(((((((.((((..((....))..))))....((((((((((....((((((....)))))).))))))..)))).)).)))))-...)).. ( -21.70) >consensus _G__GUUGCCC_AAUGGCCGCAAGGAAUUUCACACAAUUGCUAGAAAAAUUGCAGAAAAUUGAAAAUGUGCGAGAGCGCAUAAAUUUUUACUGCGUCACUUGU_AAUGCUA .......(((.....))).(((((((.((((............))))....((((((((((....((((((....)))))).))))))..)))).)).)))))........ (-22.64 = -22.25 + -0.39)

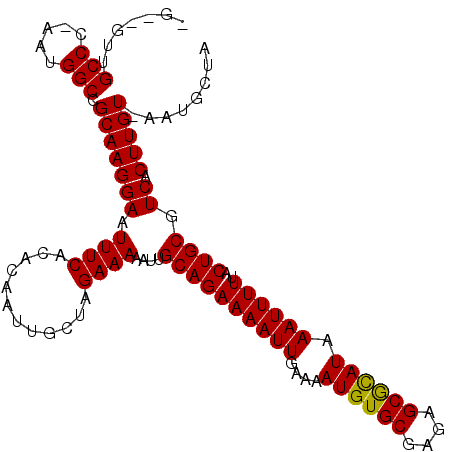
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:26 2006