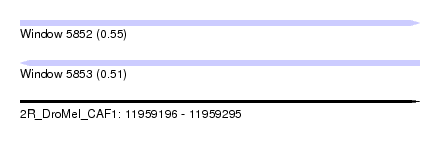
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,959,196 – 11,959,295 |
| Length | 99 |
| Max. P | 0.550895 |

| Location | 11,959,196 – 11,959,295 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -17.44 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
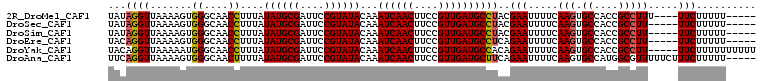
>2R_DroMel_CAF1 11959196 99 + 20766785 UAUAGGUUAAAAGUGGGCAACCUUUAUAUGCGAUUCCGUAUACAAAUCAACUUCCGUUGAUGCCUACGAAUUUUCAAGUGCCACCGCCUUU----UUCUUUUU----- ...((((.....((((((........((((((....))))))...((((((....))))))))))))((....))..........))))..----........----- ( -22.60) >DroSec_CAF1 10526 98 + 1 UAUAGGUUAAAAGUGGGCAACCUUUAUAUGCGAUUCCGUAUACAAAUCAACUUCCGUUGAUGCCUACGAAUUUUCAAGUGCCACCGCCUU-----UUCUUUUU----- ...((((.....((((((........((((((....))))))...((((((....))))))))))))((....))..........)))).-----........----- ( -22.60) >DroSim_CAF1 13671 98 + 1 UAUAGGUUAAAAGUGGGCAACCUUUAUAUGCGAUUCCGUAUACAAAUCAACUUCCGUUGAUGCCUACGAAUUUUCAAGUGCCACCGCCUU-----UUCUUUUU----- ...((((.....((((((........((((((....))))))...((((((....))))))))))))((....))..........)))).-----........----- ( -22.60) >DroEre_CAF1 10583 98 + 1 UACAGGUUAAAAGUGGGCAACCUUUAUAUGCGAUUCCGUAUACAAAUCAACUUCCGUUGAUGCCUCAGAAUUUUCAAGUGCCACCGCCUU-----UUCUUUUU----- ........(((((.((((........((((((....))))))...((((((....))))))........................)))).-----..))))).----- ( -18.30) >DroYak_CAF1 10612 103 + 1 UACAGGUUAAAAAUGGGCAACCUUUAUAUGCGAUUCCGUAUACAAAUCAACUUCCGUUGAUGCCACAGAAUUUUCAAGUGCCACCGCCUU-----UUCUUUUUUUUUU ....(((..(((((.(((........((((((....))))))...((((((....))))))))).....))))).....)))........-----............. ( -17.90) >DroAna_CAF1 11064 103 + 1 UUCAGGUUAAAAGUGGGCAACUUUUAUAUGCGAUUCCGUAUACAAAUCAACUUCCGUUGAUGCUUCAGAAUUUUCAAGUGCCAUGGCGUUUUUCUUUCUUUUU----- ...(((..(((.((.((((.(((...((((((....))))))...((((((....))))))..............)))))))...)).)))..))).......----- ( -18.20) >consensus UACAGGUUAAAAGUGGGCAACCUUUAUAUGCGAUUCCGUAUACAAAUCAACUUCCGUUGAUGCCUAAGAAUUUUCAAGUGCCACCGCCUU_____UUCUUUUU_____ ...((((.......((....))....((((((....))))))...((((((....))))))))))..(((.....(((.((....))))).....))).......... (-17.44 = -17.50 + 0.06)
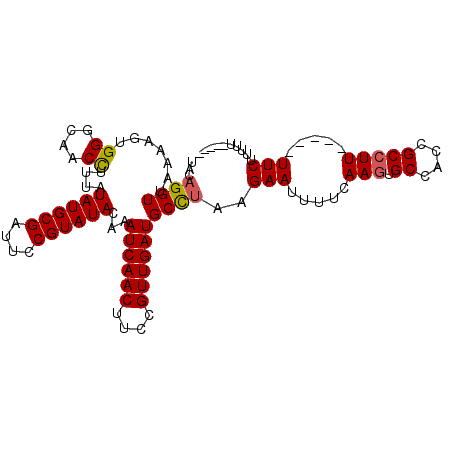
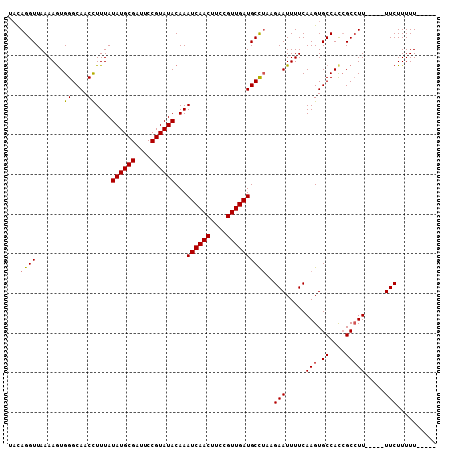
| Location | 11,959,196 – 11,959,295 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -19.72 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11959196 99 - 20766785 -----AAAAAGAA----AAAGGCGGUGGCACUUGAAAAUUCGUAGGCAUCAACGGAAGUUGAUUUGUAUACGGAAUCGCAUAUAAAGGUUGCCCACUUUUAACCUAUA -----.......(----((((..((..((.((((....((((((.((((((((....)))))..))).))))))....)).....))))..))..)))))........ ( -26.10) >DroSec_CAF1 10526 98 - 1 -----AAAAAGAA-----AAGGCGGUGGCACUUGAAAAUUCGUAGGCAUCAACGGAAGUUGAUUUGUAUACGGAAUCGCAUAUAAAGGUUGCCCACUUUUAACCUAUA -----......((-----(((..((..((.((((....((((((.((((((((....)))))..))).))))))....)).....))))..))..)))))........ ( -26.20) >DroSim_CAF1 13671 98 - 1 -----AAAAAGAA-----AAGGCGGUGGCACUUGAAAAUUCGUAGGCAUCAACGGAAGUUGAUUUGUAUACGGAAUCGCAUAUAAAGGUUGCCCACUUUUAACCUAUA -----......((-----(((..((..((.((((....((((((.((((((((....)))))..))).))))))....)).....))))..))..)))))........ ( -26.20) >DroEre_CAF1 10583 98 - 1 -----AAAAAGAA-----AAGGCGGUGGCACUUGAAAAUUCUGAGGCAUCAACGGAAGUUGAUUUGUAUACGGAAUCGCAUAUAAAGGUUGCCCACUUUUAACCUGUA -----......((-----(((..((..((.(((....((((((..((((((((....)))))..)))...))))))........)))))..))..)))))........ ( -25.90) >DroYak_CAF1 10612 103 - 1 AAAAAAAAAAGAA-----AAGGCGGUGGCACUUGAAAAUUCUGUGGCAUCAACGGAAGUUGAUUUGUAUACGGAAUCGCAUAUAAAGGUUGCCCAUUUUUAACCUGUA ..........(((-----((.(.((..((.(((....((((((((((((((((....)))))..))).))))))))........)))))..))).)))))........ ( -25.90) >DroAna_CAF1 11064 103 - 1 -----AAAAAGAAAGAAAAACGCCAUGGCACUUGAAAAUUCUGAAGCAUCAACGGAAGUUGAUUUGUAUACGGAAUCGCAUAUAAAAGUUGCCCACUUUUAACCUGAA -----................((....))...((...((((((..((((((((....)))))..)))...))))))..))..(((((((.....)))))))....... ( -19.50) >consensus _____AAAAAGAA_____AAGGCGGUGGCACUUGAAAAUUCGGAGGCAUCAACGGAAGUUGAUUUGUAUACGGAAUCGCAUAUAAAGGUUGCCCACUUUUAACCUAUA ..........(((.....(((((....)).))).....)))...(((((((((....)))))(((((((.((....)).)))))))...))))............... (-19.72 = -20.05 + 0.33)

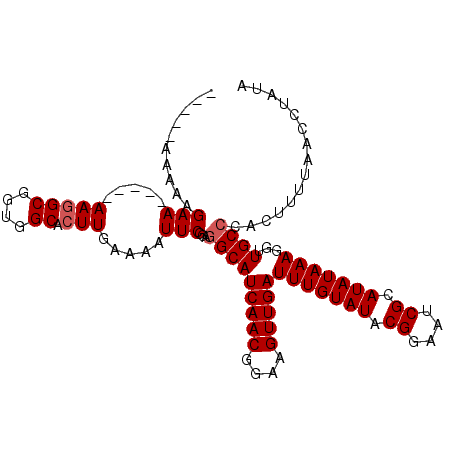
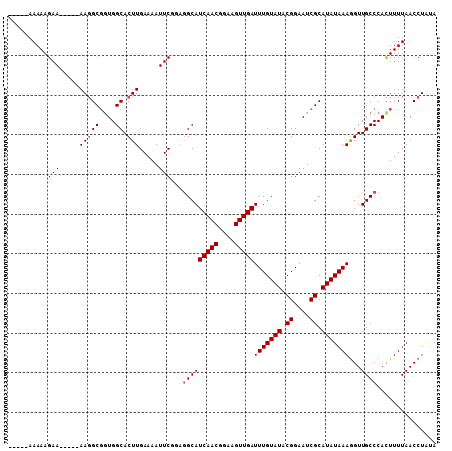
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:24 2006