| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,336,149 – 2,336,254 |
| Length | 105 |
| Max. P | 0.661605 |

| Location | 2,336,149 – 2,336,254 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -16.43 |
| Energy contribution | -17.21 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
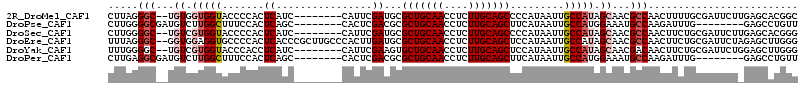
>2R_DroMel_CAF1 2336149 105 + 20766785 CUUAGGGC--UGUGGUGGUACCCCACUCAUC--------CAUUCGAUGCGCUGCAACCUCUUGCAGCCCCAUAAUUGCCAUAGCAACGCCAACUUUUGCGAUUCUUGAGCACGGC ((((((((--(((((..((........((((--------.....)))).(((((((....)))))))......))..)))))))..(((........)))...))))))...... ( -34.20) >DroPse_CAF1 108227 99 + 1 CUUGGGGCGAUGUCUUGGCUUUCCACUCAGC--------CACUCGACGCGCUGCAACCUCUUGCAGCUUCAUAAUUGCCAUGGAAAUGCCAAGAUUUG--------GAGCCUGUU ....((((...((((((((((((((....((--------........))(((((((....))))))).............)))))).))))))))...--------..))))... ( -37.10) >DroSec_CAF1 108617 105 + 1 CUUGGGGC--UGUCGUGGUACCCCACUCAUC--------CAUUCGAUGCGCUGCAACCUCUUGCAGCCCCAUAAUUGCCAUAGCAACGCCAACUUCUGCGAUUCUUGAGCACGGG .((((.(.--(((..(((((.......((((--------.....)))).(((((((....)))))))........)))))..))).).))))((..(((.........)))..)) ( -30.50) >DroEre_CAF1 100393 113 + 1 UUUAGGGC--GGUGGAGGUGCCCCACUCACCCGCUUGCCCACUUGAUGCGCUGCAACCUCUUGCAGCUCCAUAAUUGCCAUAGCAACGCCAACUUCUGCGAUUCUAGAGCUUGGG ....((((--(((((.((((...))))...)))).))))).....(((.(((((((....)))))))..)))..((((....))))..((((.(((((......))))).)))). ( -35.70) >DroYak_CAF1 97014 105 + 1 UUUGGGGC--UGUCGUGGUACCCACCUCAUC--------CAUUCGAAGUGCUGCAACCUCUUGCAGCUCCAUAAUUGCCAUAGCAACGACAACUUCUGCGAUUCUGGAGCUUGGG ....(((.--((..((((...))))..))))--------)...(.(((((((((((....)))))))((((.((((((...((.........))...)))))).)))))))).). ( -31.20) >DroPer_CAF1 110521 99 + 1 CUUGAGGCGAUGUCUUGGCUUUCCACUCAGC--------CACUCGACGCGCUGCAACCUCUUGCAGCUUCAUAAUUGCCAUGGAAAUGCCAAGAUUUG--------GAGCCUGUU ....((((...((((((((((((((....((--------........))(((((((....))))))).............)))))).))))))))...--------..))))... ( -37.10) >consensus CUUGGGGC__UGUCGUGGUACCCCACUCAUC________CACUCGAUGCGCUGCAACCUCUUGCAGCUCCAUAAUUGCCAUAGCAACGCCAACUUCUGCGAUUCU_GAGCAUGGG .....(((...((.(((((.......((................))...(((((((....))))))).........))))).))...)))......................... (-16.43 = -17.21 + 0.78)
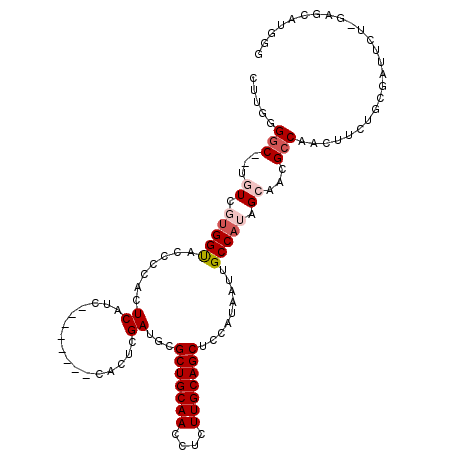
| Location | 2,336,149 – 2,336,254 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -19.39 |
| Energy contribution | -20.42 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2336149 105 - 20766785 GCCGUGCUCAAGAAUCGCAAAAGUUGGCGUUGCUAUGGCAAUUAUGGGGCUGCAAGAGGUUGCAGCGCAUCGAAUG--------GAUGAGUGGGGUACCACCACA--GCCCUAAG (((((((........(((........)))..)).)))))......((((((((((....))))))).((((.....--------)))).(((((....).)))).--.))).... ( -34.20) >DroPse_CAF1 108227 99 - 1 AACAGGCUC--------CAAAUCUUGGCAUUUCCAUGGCAAUUAUGAAGCUGCAAGAGGUUGCAGCGCGUCGAGUG--------GCUGAGUGGAAAGCCAAGACAUCGCCCCAAG ....(((..--------....(((((((.((((((((((.(((.(((.(((((((....)))))))...)))))).--------)))..))))))))))))))....)))..... ( -38.30) >DroSec_CAF1 108617 105 - 1 CCCGUGCUCAAGAAUCGCAGAAGUUGGCGUUGCUAUGGCAAUUAUGGGGCUGCAAGAGGUUGCAGCGCAUCGAAUG--------GAUGAGUGGGGUACCACGACA--GCCCCAAG ....((((..((...(((........)))...))..))))....((((((((...(.(((.((.((.((((.....--------)))).))...))))).)..))--)))))).. ( -35.30) >DroEre_CAF1 100393 113 - 1 CCCAAGCUCUAGAAUCGCAGAAGUUGGCGUUGCUAUGGCAAUUAUGGAGCUGCAAGAGGUUGCAGCGCAUCAAGUGGGCAAGCGGGUGAGUGGGGCACCUCCACC--GCCCUAAA ((((.(((.(((...(((........)))...))).)))....(((..(((((((....))))))).)))....)))).....(((((.((((((...)))))))--)))).... ( -43.30) >DroYak_CAF1 97014 105 - 1 CCCAAGCUCCAGAAUCGCAGAAGUUGUCGUUGCUAUGGCAAUUAUGGAGCUGCAAGAGGUUGCAGCACUUCGAAUG--------GAUGAGGUGGGUACCACGACA--GCCCCAAA ((((..(((((...(((.((.(((((((((....))))))))).....(((((((....))))))).)).))).))--------)).)...))))..........--........ ( -32.40) >DroPer_CAF1 110521 99 - 1 AACAGGCUC--------CAAAUCUUGGCAUUUCCAUGGCAAUUAUGAAGCUGCAAGAGGUUGCAGCGCGUCGAGUG--------GCUGAGUGGAAAGCCAAGACAUCGCCUCAAG ...((((..--------....(((((((.((((((((((.(((.(((.(((((((....)))))))...)))))).--------)))..))))))))))))))....)))).... ( -39.30) >consensus CCCAAGCUC_AGAAUCGCAAAAGUUGGCGUUGCUAUGGCAAUUAUGGAGCUGCAAGAGGUUGCAGCGCAUCGAAUG________GAUGAGUGGGGUACCACGACA__GCCCCAAG .....((((............(((((.(((....))).))))).(((.(((((((....)))))))...)))...............))))(((((...........)))))... (-19.39 = -20.42 + 1.03)

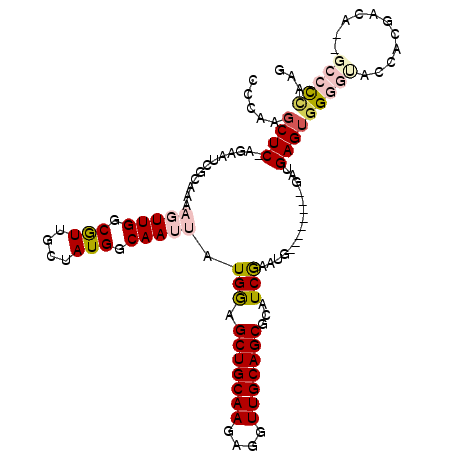
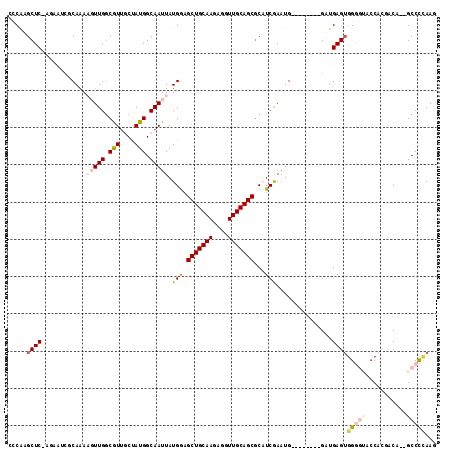
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:21 2006