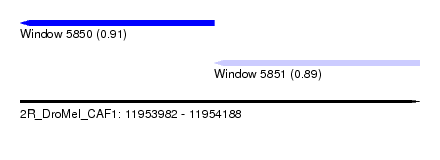
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,953,982 – 11,954,188 |
| Length | 206 |
| Max. P | 0.912332 |

| Location | 11,953,982 – 11,954,082 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -20.82 |
| Energy contribution | -22.43 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11953982 100 - 20766785 AAGCGGGCAAAUAGCGCGAUAUAAAGUUGGGGCUUUUUGGCUUUGACGCACGUAGCUCCCGAUUUGAAAGUUUGUCGUGUGUGCUU---------CGUAU---AUGUUUA-UA ..(((((((....((((((((..(((((((((((...((((......)).)).))).)))))))).......)))))))).))).)---------)))..---.......-.. ( -32.10) >DroVir_CAF1 5533 95 - 1 --------------AACGAUAUAAAGUUGGCGCUUUUUGGCUUAGACGCACGUAGCUGCCGAUUUCAAAGUUUUUCCUGUGAGUUCACGCGCGCGCGUGU---GUGUGUG-UG --------------..........((((((((((...((((......)).)).))).)))))))(((.((......)).)))...((((((((((....)---)))))))-)) ( -27.40) >DroSec_CAF1 5209 99 - 1 AAGCGGGCAUAUAGCGCGAUAUAAAGUUG-GGCUUUUUGGCUUUGACGCACGUAGCUCCCAAUUUGAAAGUUUGUCGUGUGUGCUG---------CGUAU---AUGUUUA-UA ....(((((((((.((((...((((((..-(.....)..))))))(((((((.((((..(.....)..))))...)))))))..))---------)))))---)))))).-.. ( -29.00) >DroSim_CAF1 8382 100 - 1 AAGCGGGCAAAUAGCGCGAUAUAAAGUUGGGGCUUUUUGGCUUUGACGCACGUAGCUCCCGAUUUGAAAGUUUGUCGUGUGUGCUG---------CGUAU---AUGUUUA-UA ..(((((((....((((((((..(((((((((((...((((......)).)).))).)))))))).......)))))))).)))).---------)))..---.......-.. ( -32.10) >DroYak_CAF1 5210 94 - 1 AAGCGGGCAAAUAGCGCGAUAUAAAGUUGGGGCUUUUUGGCUUUGACGCACGUAGCUCCCGAUUUGAAAGUUUGUCGUGUGUGCUC---------AGUAUACA---------- ....(((((....((((((((..(((((((((((...((((......)).)).))).)))))))).......)))))))).)))))---------........---------- ( -32.50) >DroAna_CAF1 6011 90 - 1 -----------GAGCGCGAUAUAAAGUUGGGGCUUUUUGGCUUUGACGCACGUAGCUCCCGAUUUGAAAGUUUGUCGUGUGUGCUC---------GCAAU---AUGUACACUA -----------((((((........((..(((((....)))))..))(((((.((((..(.....)..))))...)))))))))))---------.....---.......... ( -27.90) >consensus AAGCGGGCAAAUAGCGCGAUAUAAAGUUGGGGCUUUUUGGCUUUGACGCACGUAGCUCCCGAUUUGAAAGUUUGUCGUGUGUGCUC_________CGUAU___AUGUUUA_UA .....((((....((((((((..(((((((((((...((((......)).)).))).)))))))).......)))))))).))))............................ (-20.82 = -22.43 + 1.61)


| Location | 11,954,082 – 11,954,188 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 94.55 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -15.84 |
| Energy contribution | -15.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11954082 106 - 20766785 CG---GCAGCAGCAUCGAAAAGAAGGAGGGGACUUAAAAAAUAAGCUAAAAGUCUUUUGAAGUUGCCAAAGUAAAAGCAGGGGUAAAAAUG-GAACGGAGAGGUUGGGAA ..---.((((..(..((((((((......((.((((.....)))))).....))))))....(((((...((....))...))))).....-...))..)..)))).... ( -18.20) >DroSec_CAF1 5308 108 - 1 CGGUAGCAGCAGCAUCGAAAAGAAAGAGGGGACUUAAAAAAUAAGCUAAAAGUCUUUUGAAGUUGCCAAAGUAAAAGCAGGGGUAAAAAUGGGAAAG--GAGGUUGGGAA ((((.((....))))))........(((....)))..........((((...((((((..(.(((((...((....))...)))))...)..)))))--)...))))... ( -19.90) >DroSim_CAF1 8482 107 - 1 CGGCAGCAGCAGCAUCGAAAAGAAAGAGGGGACUUAAAAAAUAAGCUAAAAGUCUUUUGAAGUUGCCAAAGUAAAAGCAGGGGUAAAAAUG-GAAAG--GAGGUUGGGAA ...((((..((((.((.....))..(((....))).........)))......(((((.(..(((((...((....))...)))))...).-)))))--)..)))).... ( -19.70) >DroEre_CAF1 5335 106 - 1 CAGCGGCAGCAGCAUCGAACAGAAA-AGGGGACUUAAAAAAUAAGCUAAAAGUCUUUUGAAGUUGCCAAAGUAAAAGCAGGGGUAAAAAUG-GAGCG--GAGGUUGGGAA (((((((((((((.((.....)).(-((....))).........))).....((....)).)))))).........((.............-..)).--...)))).... ( -20.16) >DroYak_CAF1 5304 107 - 1 CAGCGGCAGCAGCAUCGAAAAGAAAGAGGGGACUUAAAAAAUAAGCUAAAAGUCUUUUGAAGUUGCCAAAGUAAAAGCAGGGGUAAAAAUG-GAACG--GAGGUUGGGAA (((((((((((((.((.....))..(((....))).........))).....((....)).))))))...((....)).............-.....--...)))).... ( -20.00) >consensus CGGCAGCAGCAGCAUCGAAAAGAAAGAGGGGACUUAAAAAAUAAGCUAAAAGUCUUUUGAAGUUGCCAAAGUAAAAGCAGGGGUAAAAAUG_GAACG__GAGGUUGGGAA ......((((..(.(((((.(((......((.((((.....)))))).....))))))))..(((((...((....))...))))).............)..)))).... (-15.84 = -15.84 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:22 2006