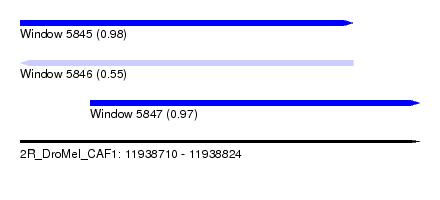
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,938,710 – 11,938,824 |
| Length | 114 |
| Max. P | 0.977921 |

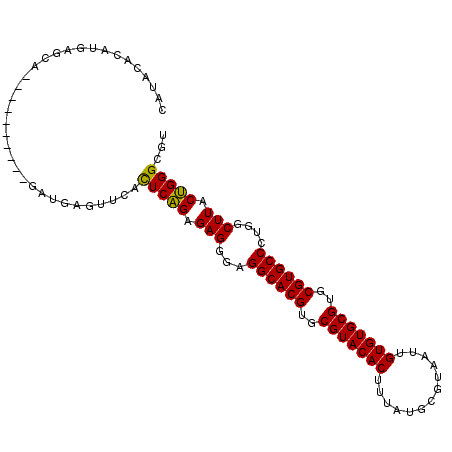
| Location | 11,938,710 – 11,938,805 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -34.15 |
| Energy contribution | -33.74 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11938710 95 + 20766785 CAUACACAUGAGCA----------GAUGAGUUCACUCAGAGAGGGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCCUGGCUUACUGGGCGU ........(((((.----------.....)))))(((((.(((((.((((((..(((((((.............)))))))..))))))))..))).)))))... ( -39.22) >DroSec_CAF1 40218 95 + 1 CAUACACAUGAGCA----------GAUGAGUUCACUCAGAGAGGGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCCUGGCUUACUGGGCGU ........(((((.----------.....)))))(((((.(((((.((((((..(((((((.............)))))))..))))))))..))).)))))... ( -39.22) >DroSim_CAF1 38075 95 + 1 CAUACACAUGAGCA----------GAUGAGUUCACUCAGAGAGGGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCCUGGCUUACUGGGCGU ........(((((.----------.....)))))(((((.(((((.((((((..(((((((.............)))))))..))))))))..))).)))))... ( -39.22) >DroEre_CAF1 37504 105 + 1 CACACACAUGAGCACAUGUACAUACAUGGGUUCACUCAGAGAGGGAGGCACGUGCGUACACUUUGUGCGUAAUUGUGUGCGUGCGUGCCAUGGCUUACUGGGCGU ........(((((.(((((....))))).)))))(((((.(((...((((((..(((((((..(.......)..)))))))..))))))....))).)))))... ( -44.30) >DroYak_CAF1 39071 88 + 1 -----------------AUACACACAUGAGUUCACUCAGAGAGGGAGGCACGUGCGUACACUUUAGGCGUAAUUGUGUGCGUGCGUGCCCUGGCUUACUGGGCGU -----------------...............(.(((((.(((((.((((((..(((((((.............)))))))..))))))))..))).))))).). ( -35.42) >DroAna_CAF1 64138 84 + 1 -------ACACACA----------AAGGA----AUUCUGGGAGGGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCUUGGCUUACGGGGCGU -------.......----------.....----.(((((..((.((((((((..(((((((.............)))))))..))))))))..))..)))))... ( -35.12) >consensus CAUACACAUGAGCA__________GAUGAGUUCACUCAGAGAGGGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCCUGGCUUACUGGGCGU ..................................(((((.(((...((((((..(((((((.............)))))))..))))))....))).)))))... (-34.15 = -33.74 + -0.42)

| Location | 11,938,710 – 11,938,805 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11938710 95 - 20766785 ACGCCCAGUAAGCCAGGGCACGCACGCACACAAUUACGCAUAAAGUGUACGCACGUGCCUCCCUCUCUGAGUGAACUCAUC----------UGCUCAUGUGUAUG ((((..((((.....(((((((..((.((((.............)))).))..))))))).......((((....))))..----------))))...))))... ( -26.22) >DroSec_CAF1 40218 95 - 1 ACGCCCAGUAAGCCAGGGCACGCACGCACACAAUUACGCAUAAAGUGUACGCACGUGCCUCCCUCUCUGAGUGAACUCAUC----------UGCUCAUGUGUAUG ((((..((((.....(((((((..((.((((.............)))).))..))))))).......((((....))))..----------))))...))))... ( -26.22) >DroSim_CAF1 38075 95 - 1 ACGCCCAGUAAGCCAGGGCACGCACGCACACAAUUACGCAUAAAGUGUACGCACGUGCCUCCCUCUCUGAGUGAACUCAUC----------UGCUCAUGUGUAUG ((((..((((.....(((((((..((.((((.............)))).))..))))))).......((((....))))..----------))))...))))... ( -26.22) >DroEre_CAF1 37504 105 - 1 ACGCCCAGUAAGCCAUGGCACGCACGCACACAAUUACGCACAAAGUGUACGCACGUGCCUCCCUCUCUGAGUGAACCCAUGUAUGUACAUGUGCUCAUGUGUGUG .......((..((....))..)).((((((((.....(((((...((((((.(((((..((.(((...))).))...))))).)))))))))))...)))))))) ( -32.10) >DroYak_CAF1 39071 88 - 1 ACGCCCAGUAAGCCAGGGCACGCACGCACACAAUUACGCCUAAAGUGUACGCACGUGCCUCCCUCUCUGAGUGAACUCAUGUGUGUAU----------------- ..((((.........))))((((((((((.....(((((.....))))).....)))).........((((....)))).))))))..----------------- ( -28.10) >DroAna_CAF1 64138 84 - 1 ACGCCCCGUAAGCCAAGGCACGCACGCACACAAUUACGCAUAAAGUGUACGCACGUGCCUCCCUCCCAGAAU----UCCUU----------UGUGUGU------- ((((..(....)...(((((((..((.((((.............)))).))..)))))))............----.....----------.))))..------- ( -18.72) >consensus ACGCCCAGUAAGCCAGGGCACGCACGCACACAAUUACGCAUAAAGUGUACGCACGUGCCUCCCUCUCUGAGUGAACUCAUC__________UGCUCAUGUGUAUG .(((.(((..((...(((((((..((.((((.............)))).))..)))))))..))..))).)))................................ (-22.10 = -22.65 + 0.56)

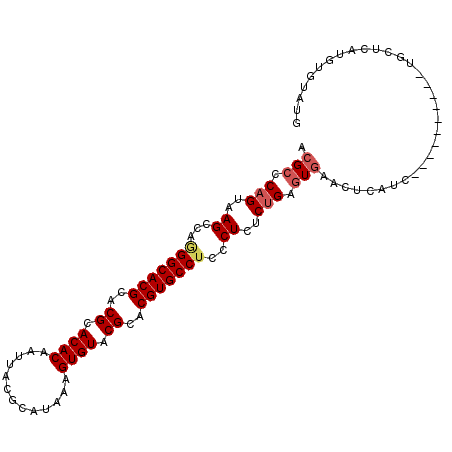
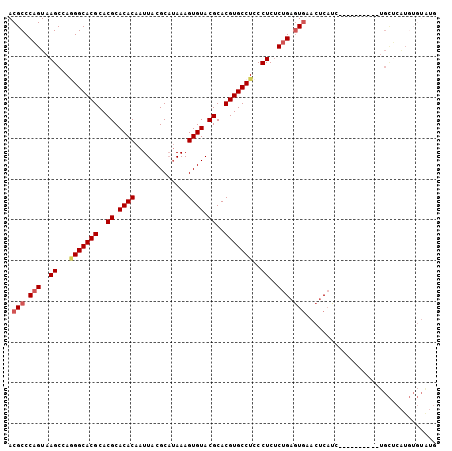
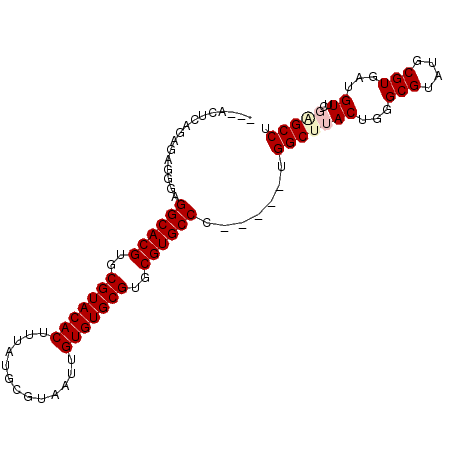
| Location | 11,938,730 – 11,938,824 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 85.66 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -34.47 |
| Energy contribution | -35.05 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11938730 94 + 20766785 UUCACUCAGAGAGGGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCC-----UGGCUUACUGGGCGUAUGCGUGAUGUUCGAGCCU ....(((...)))((.((((((..(((((((.............)))))))..)))))))-----)(((((...((((((.......))))))))))). ( -38.32) >DroPse_CAF1 45189 94 + 1 -----UAGAAGAGGGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCCAGCCACGGCGCACUGGGCGUAUGCGUGAUGUCGAGGCCU -----.........(.((((((..(((((((.............)))))))..))))))).(((.((((((((...((....))))).))))).))).. ( -40.92) >DroEre_CAF1 37534 94 + 1 UUCACUCAGAGAGGGAGGCACGUGCGUACACUUUGUGCGUAAUUGUGUGCGUGCGUGCCA-----UGGCUUACUGGGCGUAUGCGUGAUGUUCGAGCCU (((.(((...))))))((((((..(((((((..(.......)..)))))))..)))))).-----.(((((...((((((.......))))))))))). ( -39.00) >DroYak_CAF1 39084 94 + 1 UUCACUCAGAGAGGGAGGCACGUGCGUACACUUUAGGCGUAAUUGUGUGCGUGCGUGCCC-----UGGCUUACUGGGCGUAUGCGUGAUGUUCGAGCCU ....(((...)))((.((((((..(((((((.............)))))))..)))))))-----)(((((...((((((.......))))))))))). ( -38.32) >DroAna_CAF1 64150 91 + 1 ---AUUCUGGGAGGGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCU-----UGGCUUACGGGGCGUAUGCGUGAUGUUAGGGCCU ---...........((((((((..(((((((.............)))))))..)))))))-----)((((((((..(((....)))..)))..))))). ( -40.82) >DroPer_CAF1 45679 94 + 1 -----UAGAAGAGAGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCCAGCCACGGCGCACUGGGCGUAUGCGUGAUGUCGAGGCCU -----.........(.((((((..(((((((.............)))))))..))))))).(((.((((((((...((....))))).))))).))).. ( -41.12) >consensus ___ACUCAGAGAGGGAGGCACGUGCGUACACUUUAUGCGUAAUUGUGUGCGUGCGUGCCC_____UGGCUUACUGGGCGUAUGCGUGAUGUUCGAGCCU ................((((((..(((((((.............)))))))..)))))).......(((((((...(((....)))...))..))))). (-34.47 = -35.05 + 0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:19 2006