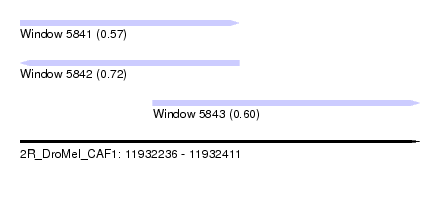
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,932,236 – 11,932,411 |
| Length | 175 |
| Max. P | 0.723292 |

| Location | 11,932,236 – 11,932,332 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.59 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.52 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
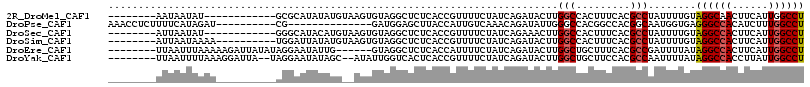
>2R_DroMel_CAF1 11932236 96 + 20766785 --------AAUAAUAU------------GCGCAUAUAUGUAAGUGUAGGCUCUCACCGUUUUCUAUCAGAUACUUGGCCACUUUCACGCCUAUUUUGUAGGCAACUUCAUUGGCCU --------........------------..((.(((((....))))).)).........................(((((.......((((((...))))))........))))). ( -18.56) >DroPse_CAF1 39175 92 + 1 AAACCUCUUUUCAUAGAU----------CG--------------GAUGGAGCUUACCAUUGUCAAACAGAUAUUGGGCCACGGCCACGGCAAUGGUGAGGGCCACAUCUUUGGCCU ....((((.(((......----------.)--------------)).))))((((((((((((...(((...)))(((....)))..))))))))))))(((((......))))). ( -35.10) >DroSec_CAF1 33774 96 + 1 --------AUUAAUAU------------GGGCAUACAUGUAAGUGUAGGCUCUCACCGUUUUCUAUCAGAAACUUGGCCACUUUCACGCCUAUUUUGUAGGCCACUUCAUUGGCCU --------..(((.((------------((((.(((((....)))))((((......(((((......)))))..))))........)))))).))).((((((......)))))) ( -26.30) >DroSim_CAF1 31689 98 + 1 --------AUUAAUAAAA----------UGGAUUAUAUGUAAGUGUAGGCUCUCACCGUUUUCUAUCAGAUACUUGGCCACUUUCACGCCUAUUUUGUAGGCCACUUCAUUGGCCU --------....((((((----------(((..(((((....)))))((((......((((......))))....))))..........)))))))))((((((......)))))) ( -19.90) >DroEre_CAF1 31042 102 + 1 --------UUAAUUUAAAAAGAUUAUAUAGGAAUAUUG------GUAGGCUCUCACCAUUUUCUAUCAGAUACUUGGCUGCUUUCACGCCGAUUUUAUAGGCCACUUCAUUGGCCU --------..........((((((..(((((((...((------((........)))).)))))))..))).)))(((((....)).)))........((((((......)))))) ( -21.60) >DroYak_CAF1 32512 104 + 1 --------UUAAUUUUAAAGGAUUA--UAGGAAUAUAGC--AUAUUGGUCACUCACCGUUUUCUAUCAGAUACUUGGCUGCUUCCACGCCAAUUUUAUAGGCCACCUUAUUGGCCU --------...............((--(((((..((((.--....((((.....))))....)))).......(((((((....)).))))))))))))(((((......))))). ( -19.10) >consensus ________AUUAAUAUAA__________AGGAAUAUAUG__AGUGUAGGCUCUCACCGUUUUCUAUCAGAUACUUGGCCACUUUCACGCCUAUUUUGUAGGCCACUUCAUUGGCCU ...........................................................................(((.........)))........((((((......)))))) ( -9.32 = -9.52 + 0.19)
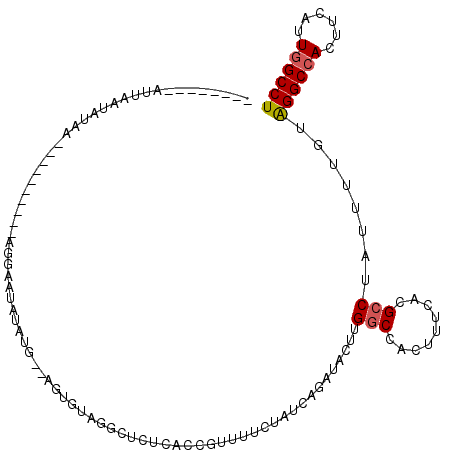
| Location | 11,932,236 – 11,932,332 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.59 |
| Mean single sequence MFE | -23.74 |
| Consensus MFE | -12.33 |
| Energy contribution | -13.17 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11932236 96 - 20766785 AGGCCAAUGAAGUUGCCUACAAAAUAGGCGUGAAAGUGGCCAAGUAUCUGAUAGAAAACGGUGAGAGCCUACACUUACAUAUAUGCGC------------AUAUUAUU-------- .(((((.......((((((.....))))))......)))))((((.((.....))....(((....)))...))))............------------........-------- ( -22.82) >DroPse_CAF1 39175 92 - 1 AGGCCAAAGAUGUGGCCCUCACCAUUGCCGUGGCCGUGGCCCAAUAUCUGUUUGACAAUGGUAAGCUCCAUC--------------CG----------AUCUAUGAAAAGAGGUUU (((((...((((..((....(((((((..(.(((....))))....((.....)))))))))..))..))))--------------..----------.(((......)))))))) ( -24.40) >DroSec_CAF1 33774 96 - 1 AGGCCAAUGAAGUGGCCUACAAAAUAGGCGUGAAAGUGGCCAAGUUUCUGAUAGAAAACGGUGAGAGCCUACACUUACAUGUAUGCCC------------AUAUUAAU-------- ((((((......))))))........((((((.(((((......((((.....))))..(((....)))..))))).....)))))).------------........-------- ( -26.80) >DroSim_CAF1 31689 98 - 1 AGGCCAAUGAAGUGGCCUACAAAAUAGGCGUGAAAGUGGCCAAGUAUCUGAUAGAAAACGGUGAGAGCCUACACUUACAUAUAAUCCA----------UUUUAUUAAU-------- .....((((((((((((((.....)))))(((.(((((........((.....))....(((....)))..))))).)))......))----------)))))))...-------- ( -24.60) >DroEre_CAF1 31042 102 - 1 AGGCCAAUGAAGUGGCCUAUAAAAUCGGCGUGAAAGCAGCCAAGUAUCUGAUAGAAAAUGGUGAGAGCCUAC------CAAUAUUCCUAUAUAAUCUUUUUAAAUUAA-------- ((((((......)))))).(((((..(((((....)).)))..((((..((((......(((....)))...------...))))...)))).....)))))......-------- ( -22.80) >DroYak_CAF1 32512 104 - 1 AGGCCAAUAAGGUGGCCUAUAAAAUUGGCGUGGAAGCAGCCAAGUAUCUGAUAGAAAACGGUGAGUGACCAAUAU--GCUAUAUUCCUA--UAAUCCUUUAAAAUUAA-------- ((((((......))))))......(((((.((....))))))).......((((.....(((.....))).....--.)))).......--.................-------- ( -21.00) >consensus AGGCCAAUGAAGUGGCCUACAAAAUAGGCGUGAAAGUGGCCAAGUAUCUGAUAGAAAACGGUGAGAGCCUACACU__CAUAUAUUCCC__________UUAUAUUAAA________ ((((((......))))))........(((.........)))..................(((....)))............................................... (-12.33 = -13.17 + 0.83)

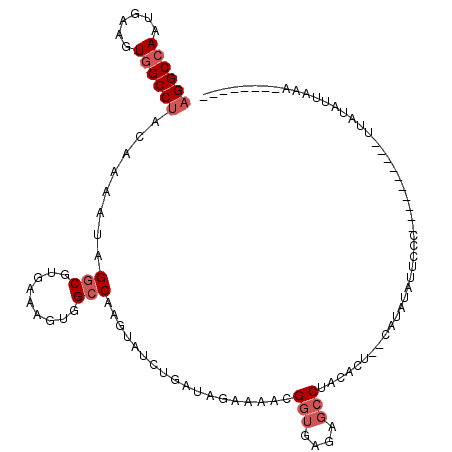

| Location | 11,932,294 – 11,932,411 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11932294 117 + 20766785 CACUUUCACGCCUAUUUUGUAGGCAACUUCAUUGGCCUGCUUCACCAGAGGAUAGAUACGUCCGCUUUGCAUCUCCUCAUUGGUUGGAAAUUCAGCGAGCUCGUGGGCGGCCAAAAG .........((((((...)))))).......((((((.(((.(((..(((((..(((.((.......)).)))))))).((.(((((....))))).))...))))))))))))... ( -34.60) >DroSec_CAF1 33832 117 + 1 CACUUUCACGCCUAUUUUGUAGGCCACUUCAUUGGCCUGCUUCACCAGAGGAUAGAUACGUCCACUCUGUAUCUCCUCAUUUGUUGGGAACUCAGCGAGCUCGUGGGCGGCUAAAAG ........(((((((...((((((((......)))))))).......(((((..((((((.......))))))))))).((((((((....))))))))...)))))))........ ( -41.40) >DroSim_CAF1 31749 117 + 1 CACUUUCACGCCUAUUUUGUAGGCCACUUCAUUGGCCUGCUUCACCAGGGGAUAGAUACGUCCACUCUGUAUCUCCUCAUUUGUUGGGAACUCAGCGAGCUCGUGGGCGGCUAAAAG ........(((((((...((((((((......)))))))).......(((((..((((((.......))))))))))).((((((((....))))))))...)))))))........ ( -40.50) >DroEre_CAF1 31106 117 + 1 UGCUUUCACGCCGAUUUUAUAGGCCACUUCAUUGGCCUGCUUCACCAGGGGAUAGAUACGUCCACUCUGUAAGUCCUCGUUUGUUGGGAAUUUAGCGAGCUCAUGGGCGCAUAACAG ........((((.((....(((((((......)))))))........((((((...((((.......)))).))))))(((((((((....)))))))))..)).))))........ ( -31.40) >DroYak_CAF1 32578 117 + 1 UGCUUCCACGCCAAUUUUAUAGGCCACCUUAUUGGCCUCCUUCACCAGGGGAUAGAUCCGUCCACUCUGUAUUUCCUCAUUAGAUGGGAACUUAGCGAGCUCAUGGGCGCACAAAAG (((.....(((.........((((((......)))))).......((((((((......)))).).)))..(((((((....)).)))))....))).(((....))))))...... ( -28.10) >DroPer_CAF1 39687 117 + 1 CACGGCCACGGCAAUGGUGAGGGCCACAUCUUUGGCCUCUGUCGCCUUUGGGUAGAUCCUGCCGCUCGCCAUCGCCUCCUCCUCGGGAUAGCCCGCCAAAGUGUGGGCGGCCACCAG ...((((..(((.(((((((((((((......))))).............(((((...))))).)))))))).)))((((....))))..((((((......))))))))))..... ( -54.60) >consensus CACUUUCACGCCUAUUUUGUAGGCCACUUCAUUGGCCUGCUUCACCAGGGGAUAGAUACGUCCACUCUGUAUCUCCUCAUUUGUUGGGAACUCAGCGAGCUCGUGGGCGGCCAAAAG ........((((......((((((((......))))))))..(((.((.((((......))))...................(((((....)))))...)).)))))))........ (-24.06 = -24.40 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:15 2006