
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,928,354 – 11,928,493 |
| Length | 139 |
| Max. P | 0.981449 |

| Location | 11,928,354 – 11,928,459 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -22.67 |
| Energy contribution | -22.75 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
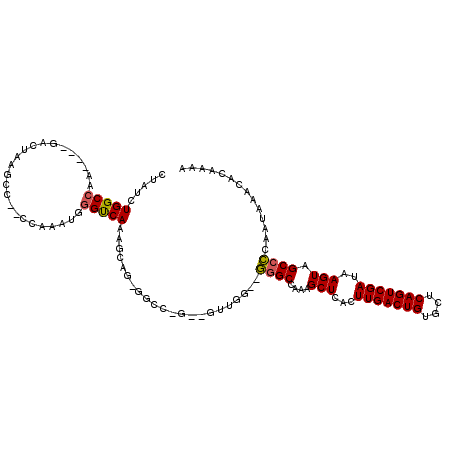
>2R_DroMel_CAF1 11928354 105 + 20766785 AAAGCAACAGAGGCCG------------AGCUGAGUAGACUUUUGUGUUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCCACCCAACUUCUGGCCUCGGCU ..(((....(((((((------------((.(((((((((......))))))).(((((((((((.(((((....)))))..)))))...))))))...)).)))..)))))).))) ( -39.30) >DroPse_CAF1 35239 114 + 1 GCAGCAACAGAAGCUGUAACCGAAGCAAGUCUGAGUAGAAUUUUGUGUUUAUUGAGGCUACUUAUCGACUGUACACAGUCAAGUGAGCUUUGGCCU--UUGACCACAGGCC-CUCCU (((((.......)))))....(((((.(((((.((((((........)))))).))))).......(((((....)))))......)))))(((((--........)))))-..... ( -34.90) >DroSec_CAF1 29942 90 + 1 AAAGCAACAGAGGCCG------------AGCUGAGUAGACUUUUGUGUUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCC--CCAAC-------------U ...........(((((------------((((.(((((((......))))))).))(((((((...(((((....))))))))).)))))))))).--.....-------------. ( -27.80) >DroYak_CAF1 28664 103 + 1 AAAGCAACAGAGGCCG------------AGCUGAGUAGACUUUUGUGAUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCC--CCAACUUGUGGCCUCGGCU ..(((....(((((((------------....((((..((....)).......((((((((((((.(((((....)))))..)))))...))))))--)..)))).))))))).))) ( -37.70) >DroAna_CAF1 54024 93 + 1 AAAGCAACAGAGGCCAA----------AAGCUGAGUAGACUUUUGUGUUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCC--CC------------CAACU ...........((((((----------(..((.(((((((......))))))).))(((((((...(((((....))))))))).)))))))))).--..------------..... ( -28.70) >DroPer_CAF1 35695 114 + 1 GCAGCAACAGAAGCCGUAACCGAAGCAAGUCUGAGUAGAAUUUUGUGUUUAUUGAGGCUACUUAUCGACUGUACACAGUCAAGUGAGCUUUGGCUU--UUGACCACAGGCC-CUCCU .......((((((((......(((((.(((((.((((((........)))))).))))).......(((((....)))))......))))))))))--))).....(((..-..))) ( -31.60) >consensus AAAGCAACAGAGGCCG____________AGCUGAGUAGACUUUUGUGUUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCC__CCAAC__C_GGCC_CGCCU ...........((((...............((.(((((((......))))))).))(((((((...(((((....))))))))).)))...))))...................... (-22.67 = -22.75 + 0.08)

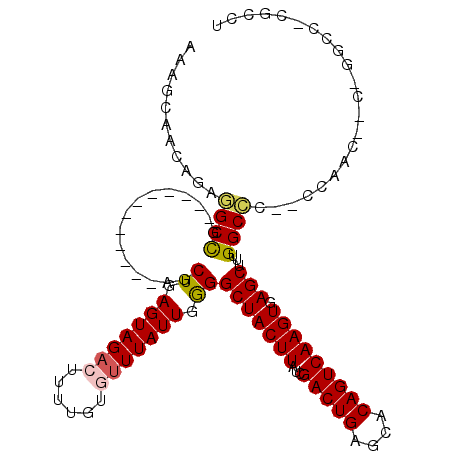
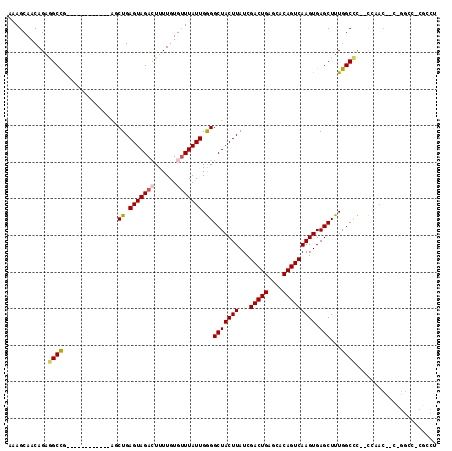
| Location | 11,928,382 – 11,928,493 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -37.99 |
| Consensus MFE | -19.92 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11928382 111 + 20766785 UUUUGUGUUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCCACCCAACUUCUGGCCUCGGCUUUGACCCAUUUGG--GGCUUAGUC----UUGGCCAGAUAG ......(((.....(((((((((((.(((((....)))))..)))))...))))))....))).(((((((..((((..(.(((....))--).)..))))----..)))))))... ( -42.80) >DroPse_CAF1 35279 108 + 1 UUUUGUGUUUAUUGAGGCUACUUAUCGACUGUACACAGUCAAGUGAGCUUUGGCCU--UUGACCACAGGCC-CUCCUCUGGCCUAUUUAA--GGCUUAGGC----UUGGCCAGAUUG ...((((.(((..((((((((((((.(((((....)))))..)))))...))))))--)))).))))((((-..(((..(((((.....)--)))).))).----..))))...... ( -39.60) >DroSec_CAF1 29970 96 + 1 UUUUGUGUUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCC--CCAAC-------------UUUGACCCAUUUGG--GUCCUAGUC----UUGGCCAGAUAG .((((.(((....((((((((((((.(((((....)))))..)))))...))))))--)..((-------------(..(((((....))--)))..))).----..)))))))... ( -32.90) >DroYak_CAF1 28692 113 + 1 UUUUGUGAUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCC--CCAACUUGUGGCCUCGGCUUUGACUCAUUUGG--AGCUUAGACUUACUUGGCCAGAUAG .............((((((((((((.(((((....)))))..)))))...))))))--).....(.(((((..(((((..(.....)..)--))))...........))))).)... ( -33.92) >DroAna_CAF1 54054 99 + 1 UUUUGUGUUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCC--CC------------CAACUUUGACCCAUUUGGGGGGCCAAGUC----UCGGCCAAAUGG .........((((..((((.......(((((....)))))....(((.((((((((--((------------(((...........))))))))))))).)----)))))).)))). ( -41.80) >DroPer_CAF1 35735 108 + 1 UUUUGUGUUUAUUGAGGCUACUUAUCGACUGUACACAGUCAAGUGAGCUUUGGCUU--UUGACCACAGGCC-CUCCUCUGGCCUAUUUAA--GGCUUAGGC----UUGGCCAGAUUG ...((((.(((..((((((((((((.(((((....)))))..)))))...))))))--)))).))))((((-..(((..(((((.....)--)))).))).----..))))...... ( -36.90) >consensus UUUUGUGUUUAUUGGGGCUACUUAUCGACUGAGCACAGUCAAGUGAGCUUUGGCCC__CCAAC__C_GGCC_CGCCUUUGACCCAUUUGG__GGCUUAGUC____UUGGCCAGAUAG ..............(((((((((((.(((((....)))))..)))))...))))))....................................((((...........))))...... (-19.92 = -19.45 + -0.47)

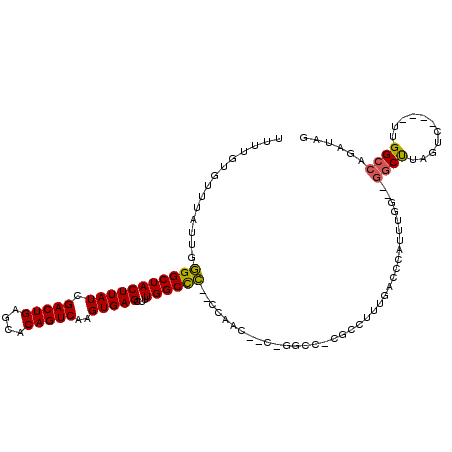
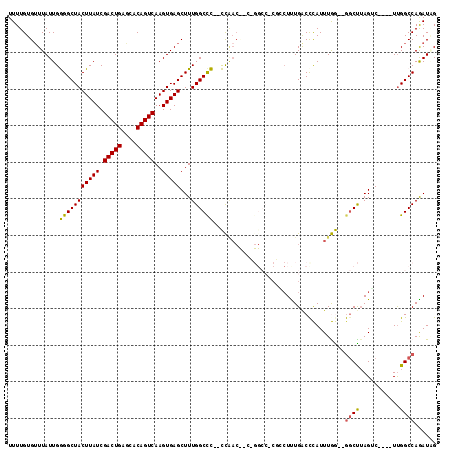
| Location | 11,928,382 – 11,928,493 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -17.83 |
| Energy contribution | -17.49 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11928382 111 - 20766785 CUAUCUGGCCAA----GACUAAGCC--CCAAAUGGGUCAAAGCCGAGGCCAGAAGUUGGGUGGGCCAAAGCUCACUUGACUGUGCUCAGUCGAUAAGUAGCCCCAAUAAACACAAAA ...(((((((..----(.((..(((--(.....))))...)).)..))))))).(((((((((((....))))))(((((((....))))))).........))))).......... ( -39.90) >DroPse_CAF1 35279 108 - 1 CAAUCUGGCCAA----GCCUAAGCC--UUAAAUAGGCCAGAGGAG-GGCCUGUGGUCAA--AGGCCAAAGCUCACUUGACUGUGUACAGUCGAUAAGUAGCCUCAAUAAACACAAAA ......((((..----.(((..(((--(.....))))...)))..-))))((((.....--((((....(((...(((((((....)))))))..))).)))).......))))... ( -35.60) >DroSec_CAF1 29970 96 - 1 CUAUCUGGCCAA----GACUAGGAC--CCAAAUGGGUCAAA-------------GUUGG--GGGCCAAAGCUCACUUGACUGUGCUCAGUCGAUAAGUAGCCCCAAUAAACACAAAA .....(((((..----((((..(((--((....)))))..)-------------)))..--.)))))..(((.(((((((((....)))))...)))))))................ ( -33.90) >DroYak_CAF1 28692 113 - 1 CUAUCUGGCCAAGUAAGUCUAAGCU--CCAAAUGAGUCAAAGCCGAGGCCACAAGUUGG--GGGCCAAAGCUCACUUGACUGUGCUCAGUCGAUAAGUAGCCCCAAUAAUCACAAAA .....(((((.((.....))..(((--(.....)))).........))))).......(--((((....(((...(((((((....)))))))..))).)))))............. ( -29.40) >DroAna_CAF1 54054 99 - 1 CCAUUUGGCCGA----GACUUGGCCCCCCAAAUGGGUCAAAGUUG------------GG--GGGCCAAAGCUCACUUGACUGUGCUCAGUCGAUAAGUAGCCCCAAUAAACACAAAA ......(((.((----(..((((((((((((.(.......).)))------------))--)))))))..)))..(((((((....)))))))......)))............... ( -40.40) >DroPer_CAF1 35735 108 - 1 CAAUCUGGCCAA----GCCUAAGCC--UUAAAUAGGCCAGAGGAG-GGCCUGUGGUCAA--AAGCCAAAGCUCACUUGACUGUGUACAGUCGAUAAGUAGCCUCAAUAAACACAAAA ......((((..----.(((..(((--(.....))))...)))..-))))..((((...--..))))..(((.(((((((((....)))))...)))))))................ ( -32.20) >consensus CUAUCUGGCCAA____GACUAAGCC__CCAAAUGGGUCAAAGCAG_GGCC_G__GUUGG__GGGCCAAAGCUCACUUGACUGUGCUCAGUCGAUAAGUAGCCCCAAUAAACACAAAA .....(((((........................)))))......................((((....(((...(((((((....)))))))..))).)))).............. (-17.83 = -17.49 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:11 2006