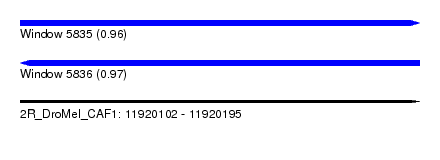
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,920,102 – 11,920,195 |
| Length | 93 |
| Max. P | 0.969018 |

| Location | 11,920,102 – 11,920,195 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -27.62 |
| Energy contribution | -28.46 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11920102 93 + 20766785 CUCUGCGGUGAGCAGAGGUGUG----UUUGCUGG-GGGAAAAGGGACGAUGGCGACGCGCCUUGUUUGGCCCGGCAAAUGUGGCUUAAUAGCAAAAUA ((((((.....))))))..(..----((((((((-(..(((((((.((.((....)))))))).)))..)))))))))..).(((....)))...... ( -36.50) >DroSec_CAF1 21794 96 + 1 CUCUGCGGUGAGCAGAGGUGUGUGUGUUUGCUGGGGGGGAAAGGGACGC--GCGACGCGCCUUGUUUGGCCCGGCAAAUGUGGCUUAAUAGCAAAAUA ((((((.....))))))......(..(((((((((..(((.((((.(((--(...)))))))).)))..)))))))))..).(((....)))...... ( -39.40) >DroSim_CAF1 19709 98 + 1 CUCUGCGGUGAGCAGAGGUGUGUUUGUUUGCUGGGGGGAAAAGGGACGAUGGCGACGCGCCUUGUUUGGCCCGGCAAAUGCGGCUUAAUAGCAAAAUA ((((((.....))))))...(((..((((((((((..(((.((((.((.((....)))))))).)))..)))))))))))))(((....)))...... ( -37.60) >DroEre_CAF1 18918 94 + 1 CUUUGCGGGGGGCAGAGGUGUG----UUUGCCAGGGGGAAAAGGGACGAUGGCGACGCGCCUUGUUUGGCCCGGCAAAUAUGGCUUAAUAGCUAAAUA .(((((.(((((((.(((((((----((.((((..(..........)..)))))))))))))))))...))).)))))..(((((....))))).... ( -36.90) >DroYak_CAF1 20050 93 + 1 CUUUGCGGUGAGCAGCGGUGUG----UUUGCU-GGGGGAAAAGGGACAAUGGCGACGCGCCUUGUUUGGCCCGGCAAAUGUGGCUUAAUAGCAAAAUA .(((((..(((((.((.....(----((((((-(((.......((((((.((((...))))))))))..)))))))))))).)))))...)))))... ( -33.50) >consensus CUCUGCGGUGAGCAGAGGUGUG____UUUGCUGGGGGGAAAAGGGACGAUGGCGACGCGCCUUGUUUGGCCCGGCAAAUGUGGCUUAAUAGCAAAAUA ((((((.....)))))).........(((((((((..(((.((((.((.((....)))))))).)))..)))))))))....(((....)))...... (-27.62 = -28.46 + 0.84)

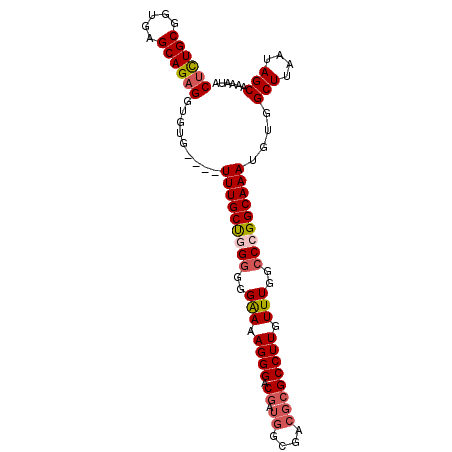
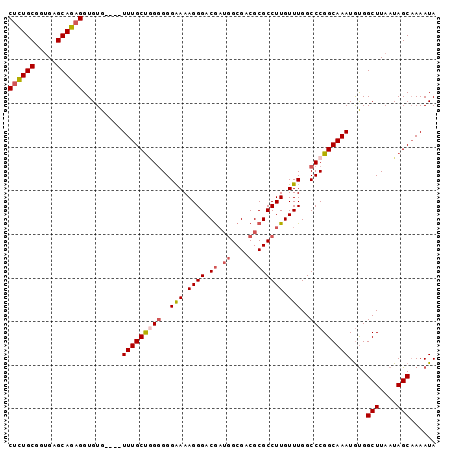
| Location | 11,920,102 – 11,920,195 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -19.64 |
| Energy contribution | -20.36 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11920102 93 - 20766785 UAUUUUGCUAUUAAGCCACAUUUGCCGGGCCAAACAAGGCGCGUCGCCAUCGUCCCUUUUCCC-CCAGCAAA----CACACCUCUGCUCACCGCAGAG ......(((....)))....(((((.(((......((((.(((.......))).))))....)-)).)))))----.....((((((.....)))))) ( -24.70) >DroSec_CAF1 21794 96 - 1 UAUUUUGCUAUUAAGCCACAUUUGCCGGGCCAAACAAGGCGCGUCGC--GCGUCCCUUUCCCCCCCAGCAAACACACACACCUCUGCUCACCGCAGAG ......(((....)))....(((((.(((........((((((...)--))))).........))).))))).........((((((.....)))))) ( -26.13) >DroSim_CAF1 19709 98 - 1 UAUUUUGCUAUUAAGCCGCAUUUGCCGGGCCAAACAAGGCGCGUCGCCAUCGUCCCUUUUCCCCCCAGCAAACAAACACACCUCUGCUCACCGCAGAG ......(((....)))....(((((.(((......((((.(((.......))).)))).....))).))))).........((((((.....)))))) ( -25.10) >DroEre_CAF1 18918 94 - 1 UAUUUAGCUAUUAAGCCAUAUUUGCCGGGCCAAACAAGGCGCGUCGCCAUCGUCCCUUUUCCCCCUGGCAAA----CACACCUCUGCCCCCCGCAAAG ......(((....)))....(((((((((......((((.(((.......))).)))).....)))))))))----.....((.(((.....))).)) ( -22.50) >DroYak_CAF1 20050 93 - 1 UAUUUUGCUAUUAAGCCACAUUUGCCGGGCCAAACAAGGCGCGUCGCCAUUGUCCCUUUUCCCCC-AGCAAA----CACACCGCUGCUCACCGCAAAG ...(((((.....(((....(((((.(((....((((((((...)))).)))).........)))-.)))))----......))).......))))). ( -20.22) >consensus UAUUUUGCUAUUAAGCCACAUUUGCCGGGCCAAACAAGGCGCGUCGCCAUCGUCCCUUUUCCCCCCAGCAAA____CACACCUCUGCUCACCGCAGAG ......(((....)))....(((((.(((......((((.(((.......))).)))).....))).))))).........((((((.....)))))) (-19.64 = -20.36 + 0.72)

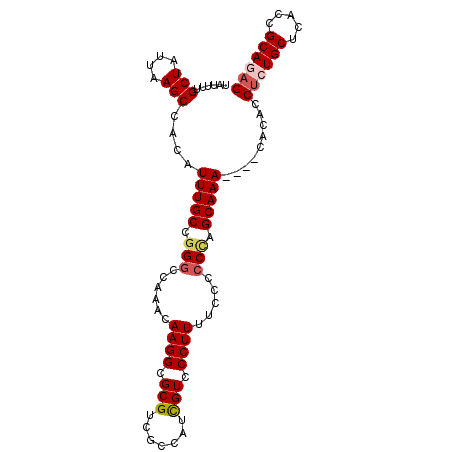
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:08 2006