
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,897,520 – 11,897,816 |
| Length | 296 |
| Max. P | 0.973818 |

| Location | 11,897,520 – 11,897,637 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.47 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11897520 117 + 20766785 UUAACUAUAAAUUCUGUGUGGCAAAACAUGCAAACAUGUUUGCGGAAAUACCUGACAGACUCAGGAGGGUCAAACCAAGGGGGGGGGGGGUGUGGUUCGGUGCAGGAAAGAUCA---GGG ...........(((((..(.(((((.((((....)))))))))(((.((((((..(.(((((....)))))...((......)))..))))))..))).)..))))).......---... ( -32.90) >DroSec_CAF1 38008 110 + 1 UUAACUAUAAAUUCUGUGUGGCAAAACAUGGAAAAAUGUUUGCGGAAAUACCUGACAGACUCAGGAGGGUCAAACCAGGGGGUGGGG------GUUUCGGUGCAGA-AAGAUCA---GGU ...........(((((..(.(((((.(((......)))))))).(((((.(((.((.(((((....)))))...((...)))).)))------))))).)..))))-)......---... ( -29.70) >DroSim_CAF1 38412 116 + 1 UUAACUAUAAAUUCUGUGUGGCAAAACAUGCAAACAUGUUUGCGGAAAUACCUGACAGACUCAGGAGGGUCAAUCCAGGGGGGGAGG-GGGGUGGUUUGCUGCAGAAAAGAUCA---GGU ...........((((((..((((((...((((((....))))))....(((((..(.(((((....)))))..(((......)))).-.))))).)))))))))))).......---... ( -32.60) >DroYak_CAF1 38559 115 + 1 UUAACUAUAAAUUCUGUGUGGCAAAACAUGCAAACAUGUUUGCGGAAAUACCUGACAGACUCGGGAGAGUCAACCUAGGGCGGGAGGAGGUG-GGUU-C---CUGAAAAGAUUAAAGGGG ....((......(((.....(((((.((((....)))))))))((((.(((((....(((((....)))))..(((........))))))))-..))-)---).....)))......)). ( -32.10) >consensus UUAACUAUAAAUUCUGUGUGGCAAAACAUGCAAACAUGUUUGCGGAAAUACCUGACAGACUCAGGAGGGUCAAACCAGGGGGGGAGG_GGUG_GGUUCGGUGCAGAAAAGAUCA___GGG ...........((((((...(((((.((((....))))))))).......((((...(((((....)))))....))))......................))))))............. (-20.26 = -20.95 + 0.69)

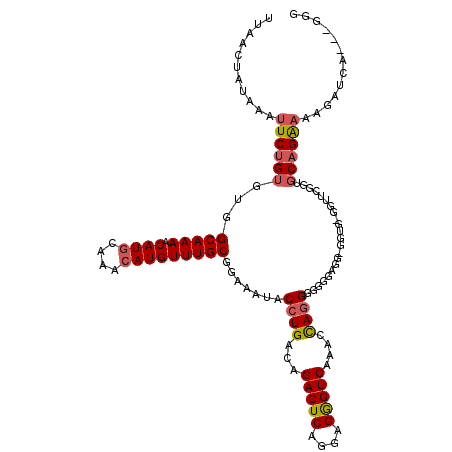

| Location | 11,897,520 – 11,897,637 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.47 |
| Mean single sequence MFE | -21.99 |
| Consensus MFE | -15.09 |
| Energy contribution | -16.15 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11897520 117 - 20766785 CCC---UGAUCUUUCCUGCACCGAACCACACCCCCCCCCCCCUUGGUUUGACCCUCCUGAGUCUGUCAGGUAUUUCCGCAAACAUGUUUGCAUGUUUUGCCACACAGAAUUUAUAGUUAA ..(---((......((((((..((((((...............))))))(((.(....).))))).)))).......(((((((((....)))).)))))....)))............. ( -22.16) >DroSec_CAF1 38008 110 - 1 ACC---UGAUCUU-UCUGCACCGAAAC------CCCCACCCCCUGGUUUGACCCUCCUGAGUCUGUCAGGUAUUUCCGCAAACAUUUUUCCAUGUUUUGCCACACAGAAUUUAUAGUUAA ..(---((....(-((((.........------........(((((...(((.(....).)))..))))).......((((((((......))).)))))....)))))....))).... ( -17.80) >DroSim_CAF1 38412 116 - 1 ACC---UGAUCUUUUCUGCAGCAAACCACCCC-CCUCCCCCCCUGGAUUGACCCUCCUGAGUCUGUCAGGUAUUUCCGCAAACAUGUUUGCAUGUUUUGCCACACAGAAUUUAUAGUUAA ..(---((.....(((((..(((((.((....-........(((((...(((.(....).)))..))))).......(((((....))))).)).)))))....)))))....))).... ( -23.50) >DroYak_CAF1 38559 115 - 1 CCCCUUUAAUCUUUUCAG---G-AACC-CACCUCCUCCCGCCCUAGGUUGACUCUCCCGAGUCUGUCAGGUAUUUCCGCAAACAUGUUUGCAUGUUUUGCCACACAGAAUUUAUAGUUAA ...((.(((....(((.(---(-((..-.((((....((......))..(((((....)))))....))))..))))(((((((((....)))).)))))......))).))).)).... ( -24.50) >consensus ACC___UGAUCUUUUCUGCACCGAACC_CACC_CCCCCCCCCCUGGUUUGACCCUCCUGAGUCUGUCAGGUAUUUCCGCAAACAUGUUUGCAUGUUUUGCCACACAGAAUUUAUAGUUAA .............(((((.......................(((((...(((.(....).)))..))))).......(((((((((....)))).)))))....)))))........... (-15.09 = -16.15 + 1.06)

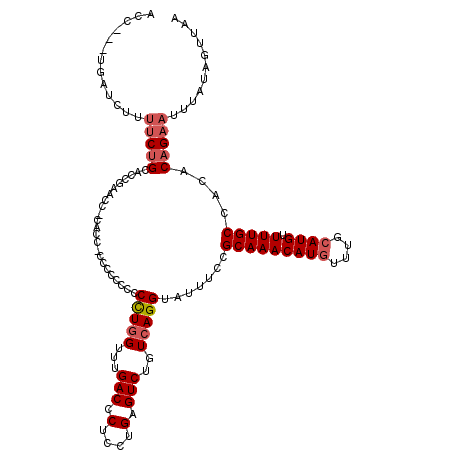
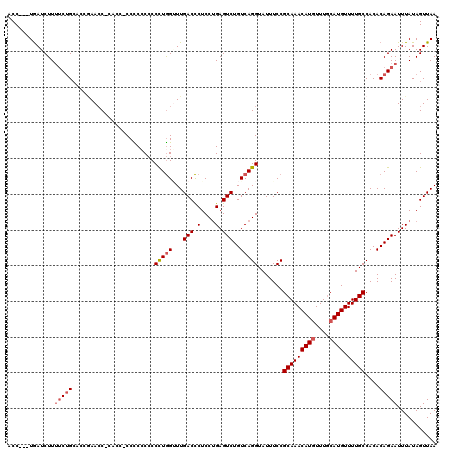
| Location | 11,897,600 – 11,897,716 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -44.59 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.68 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11897600 116 + 20766785 GGGGGGGGGGUGUGGUUCGGUGCAGGAAAGAUCA---GGGGAGGUGGCUGGGGAGUGGCACAAUCGAGCAGCCGAGGUGUCAAGCGUUGCAAACACCUUGGUGACAUCGAACC-CUCCGC ((((((..(((((.(((((((((........(((---(.........))))......))))...))))).((((((((((...((...))..)))))))))).)))))...))-)))).. ( -45.24) >DroSec_CAF1 38088 109 + 1 GGUGGGG------GUUUCGGUGCAGA-AAGAUCA---GGUGAGGUGGCAGGGGAGGGGCACAAUCGAGCAGCCGAGGUGUCAAGCGUUGCAAACACCUUGGUGACAUCGAACC-CUCCAC .((((((------(.((((((((...-.......---(((...(((.(........).))).))).....((((((((((...((...))..))))))))))).))))))).)-)))))) ( -42.20) >DroSim_CAF1 38492 115 + 1 GGGGAGG-GGGGUGGUUUGCUGCAGAAAAGAUCA---GGUGAGGUGGCAGGGGAGUGGCACAAUCGAGCAGCCGAGGUGUCAAGCGUUGCAAACACCUUGGUGACAUCGAACC-CUCCAC ..(((((-(.(.((.(((.((((........((.---...))....)))).))).)).)....((((...((((((((((...((...))..))))))))))....)))).))-)))).. ( -42.90) >DroYak_CAF1 38639 114 + 1 CGGGAGGAGGUG-GGUU-C---CUGAAAAGAUUAAAGGGGGGGGUGGCG-GGGGGUGCCACAAUCGAGCAGCCGAGGUGUCAAGCGUUGCAAACACCUUGGUGACAUCGAACCCCUCGAC ((((((......-..))-)---)))...........((((((.((((((-.....))))))..((((...((((((((((...((...))..))))))))))....)))).))))))... ( -48.00) >consensus GGGGAGG_GGUG_GGUUCGGUGCAGAAAAGAUCA___GGGGAGGUGGCAGGGGAGUGGCACAAUCGAGCAGCCGAGGUGUCAAGCGUUGCAAACACCUUGGUGACAUCGAACC_CUCCAC ((((.((....................................(((.(........).)))..((((...((((((((((...((...))..))))))))))....)))).)).)))).. (-30.80 = -30.68 + -0.12)

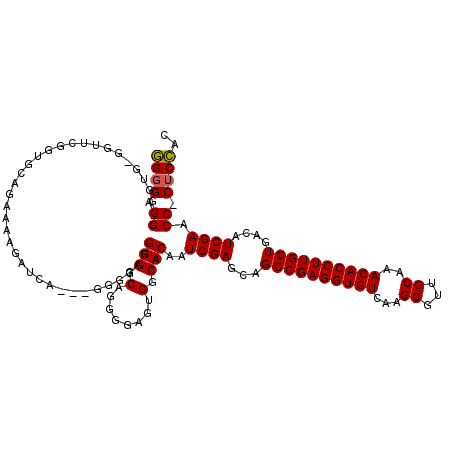
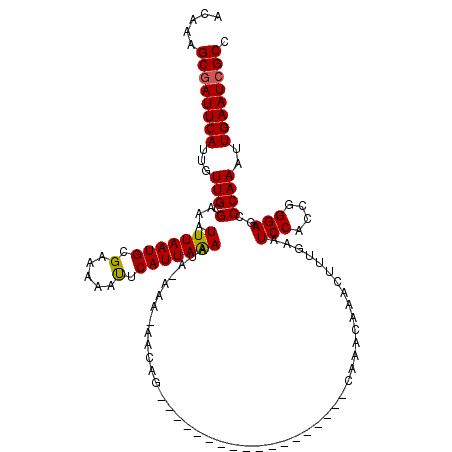
| Location | 11,897,716 – 11,897,816 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.72 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.08 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11897716 100 + 20766785 ACAAAGCGAUUCAUUGUUGCGAAAUUUAAUGCGAAAAAUUCAUUAAAAUA-AAA-AACAG------------------CAAACAAACUUUGAAUCCACCGGGAGCCCAAAUUGAAUCGCC .....((((((((((((((.....(((((((.........)))))))...-...-..)))------------------)))......((((..(((....)))...)))).)))))))). ( -20.22) >DroSec_CAF1 38197 99 + 1 ACAAAGCGAUUCAUUGUUGCGAAAUUUAAUGCGAAAAACUCAUUAAAAUA-AAA-AA-AG------------------CAAACAAACUUUGAAUCCACCGGGAACCCAAAUUGAAUCGCC .....(((((((((((((((....(((((((.(.....).)))))))...-...-..-.)------------------).)))))..............((....))....)))))))). ( -20.22) >DroSim_CAF1 38607 100 + 1 ACAAAGCGAUUCAUUGUUGCGAAAUUUAAUGCGAAAAACUCAUUAAAAUA-AAA-AACAG------------------CAAACAAACUUUGAAUCCACCGGGAGCCCAAAUUGAAUCGCC .....((((((((((((((.....(((((((.(.....).)))))))...-...-..)))------------------)))......((((..(((....)))...)))).)))))))). ( -21.62) >DroEre_CAF1 40151 119 + 1 ACAAAGCGAUUCAUUGUUGCGAAAUUUAAUGCGAAAAAUUCAUUAGAAUACAAAAAACAGCUCGGCAGGAGGAAUUGCCAAACAAACUUUGAAUCCACCGGGAGCCCAAAUUGAAU-GCC .....((((((((..(((......(((((((.........)))))))................(((((......))))).....)))..))))))....((....)).........-)). ( -21.90) >DroYak_CAF1 38753 119 + 1 ACAAAGCGAUUCAUUGUUGCGAAAUUUAAUGCGAAAAAUUCAUUAAAAUA-AAAAAACACCUCUGCAUGAAGAAUUGCCAAACAAACUUUGAAUCCACCGGGAGUCCAAAUUGAAUCGCC .....(((((((((((((((((..(((((((.........)))))))...-..........(((......))).))))..)))))..((((..(((....)))...)))).)))))))). ( -23.40) >consensus ACAAAGCGAUUCAUUGUUGCGAAAUUUAAUGCGAAAAAUUCAUUAAAAUA_AAA_AACAG__________________CAAACAAACUUUGAAUCCACCGGGAGCCCAAAUUGAAUCGCC .....((((((((...(((.(...(((((((.(.....).)))))))..............................................(((....)))..))))..)))))))). (-16.28 = -16.08 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:00 2006