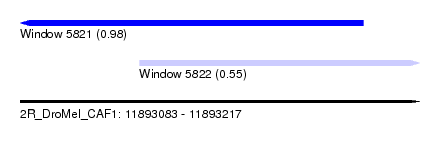
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,893,083 – 11,893,217 |
| Length | 134 |
| Max. P | 0.984912 |

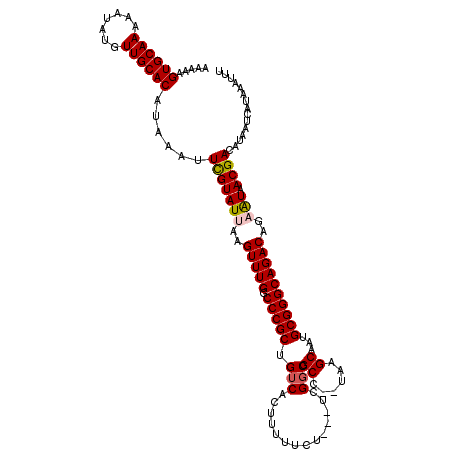
| Location | 11,893,083 – 11,893,198 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -25.05 |
| Energy contribution | -25.19 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11893083 115 - 20766785 AAAAAGUGCAAAAAUAUGUUGCACAUAAAUUCGUAUUAAGUUUGGCCCGCUGUCACUUUUUCU---UCGGCCGCC--UAAGCAAUGCGGGCAGACAGAAUAACGACAUAAUCAUAAAUUU .....((((((.......))))))......(((((((..(((((.(((((...........(.---...)..((.--...))...))))))))))..))).))))............... ( -28.00) >DroPse_CAF1 31669 120 - 1 AAAAAGUGCAAAAAUAUGUUGCACAUAAAUUUGUAUUAAGUUUGGCCCGCUGUCACUUUUUCUUCUUCGGCCGCCUCUAAGCAAUGCGGGCAGACAGCAUAACGACAUAAUCAUAAAUUU .....((((((.......))))))..((((((((((((.(((.(....((((((..........(....)..((((...(....)..)))).))))))....)))).)))).)))))))) ( -27.80) >DroSec_CAF1 33605 115 - 1 AAAAAGUGCAAAAAUAUGUUGCACAUAAAUUCGUAUUAAGUUUGGCCCGCUGUCACUUUUUCU---UCGGCCGCC--UAAGCAAUGCGGGCAGACAGAAUAACGACAUAAUCAUAAAUUU .....((((((.......))))))......(((((((..(((((.(((((...........(.---...)..((.--...))...))))))))))..))).))))............... ( -28.00) >DroSim_CAF1 33954 113 - 1 AAAAAGUGCAAAAAUAUGUUGCACAUAAAUUCGUAUUAAGUUUGGCCCGCUGUCACUUUUUCU---UCGGCCGCC--UAAGC-AUGCGGGCAGACAGAAUAACGACAUA-UCAUAAAUUU .....((((((.......))))))......(((((((..(((((.((((((((........(.---...).....--...))-).))))))))))..))).))))....-.......... ( -29.29) >DroEre_CAF1 35540 114 - 1 AGAAAGUGCAAAAAUAUGUUGCACAUAAAUUCGUAUUAAGUUUGGCCCGCUGUCACUUUUUCU---UCG-CCGCC--UAAGCAAUGCGGGCAGACAGAGUAACGACAUAAUCAUAAAUUU .((..((((((.......))))))......(((((((..(((((.(((((.....(((...(.---...-..)..--.)))....))))))))))..))).)))).....))........ ( -26.00) >DroPer_CAF1 32995 120 - 1 AAAAAGUGCAAAAAUAUGUUGCACAUAAAUUUGUAUUAAGUUUGGCCCGCUGUCACUUUUUCUUCUUCGGCCGCCUCUAAGCAAUGCGGGCAGACAGCAUAACGACAUAAUCAUAAAUUU .....((((((.......))))))..((((((((((((.(((.(....((((((..........(....)..((((...(....)..)))).))))))....)))).)))).)))))))) ( -27.80) >consensus AAAAAGUGCAAAAAUAUGUUGCACAUAAAUUCGUAUUAAGUUUGGCCCGCUGUCACUUUUUCU___UCGGCCGCC__UAAGCAAUGCGGGCAGACAGAAUAACGACAUAAUCAUAAAUUU .....((((((.......))))))......(((((((..(((((.(((((.(((..............))).((......))...))))))))))..))).))))............... (-25.05 = -25.19 + 0.14)

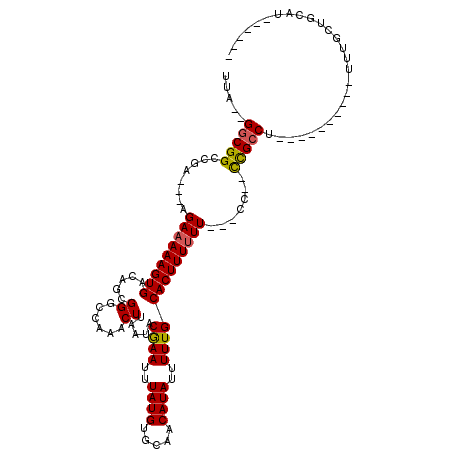
| Location | 11,893,123 – 11,893,217 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -19.42 |
| Energy contribution | -19.09 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11893123 94 + 20766785 UUA--GGCGGCCGA---AGAAAAAGUGACAGCGGGCCAAACUUAAUACGAAUUUAUGUGCAACAUAUUUUUGCACUUUUUU---CC--CCGCCC----------UUUGAAGCAU------ ...--(((((....---.(((((((((.....((......)).....((((..((((.....))))..)))))))))))))---..--))))).----------..........------ ( -20.70) >DroPse_CAF1 31709 117 + 1 UUAGAGGCGGCCGAAGAAGAAAAAGUGACAGCGGGCCAAACUUAAUACAAAUUUAUGUGCAACAUAUUUUUGCACUUUUUU---UUUGUUGCCUUUGGUGUGCCUUUGGUGUAUGCCCAG ..(((((((((....((((((((((((.....((......)).....((((..((((.....))))..)))))))))))))---))))))))))))((((((((....).)))))))... ( -33.10) >DroSim_CAF1 33992 94 + 1 UUA--GGCGGCCGA---AGAAAAAGUGACAGCGGGCCAAACUUAAUACGAAUUUAUGUGCAACAUAUUUUUGCACUUUUUU---CC--CCGGCG----------UUUGCAGCAU------ .((--((((.(((.---.(((((((((.....((......)).....((((..((((.....))))..)))))))))))))---..--.)))))----------))))......------ ( -21.20) >DroEre_CAF1 35580 93 + 1 UUA--GGCGG-CGA---AGAAAAAGUGACAGCGGGCCAAACUUAAUACGAAUUUAUGUGCAACAUAUUUUUGCACUUUCUU---CC--CCGCCA----------UUUGCUGCAC------ ...--(((((-.((---(((((..((((...((..............))...))))((((((.......))))))))))))---).--))))).----------..........------ ( -25.94) >DroYak_CAF1 34083 97 + 1 UUA--GGCGGCCGA---AGAAAAAGUGACAGCGGGCCAAACUUAAUACGAAUUUAUGUGCAACAUAUUUUUGCACUUUUUUUCGCC--CCGCCU----------UUUGCUGCAU------ ...--.(((((.((---((...........(((((............((((.....((((((.......)))))).....)))).)--))))))----------)).)))))..------ ( -25.72) >DroPer_CAF1 33035 117 + 1 UUAGAGGCGGCCGAAGAAGAAAAAGUGACAGCGGGCCAAACUUAAUACAAAUUUAUGUGCAACAUAUUUUUGCACUUUUUU---UUUGUUGCCUUUGGUGUGCCUUUGGUGUGUGCCCAG ..(((((((((....((((((((((((.....((......)).....((((..((((.....))))..)))))))))))))---))))))))))))((((..((....).)..))))... ( -31.70) >consensus UUA__GGCGGCCGA___AGAAAAAGUGACAGCGGGCCAAACUUAAUACGAAUUUAUGUGCAACAUAUUUUUGCACUUUUUU___CC__CCGCCU__________UUUGCUGCAU______ .....(((((........(((((((((.....((......)).....((((..((((.....))))..))))))))))))).......)))))........................... (-19.42 = -19.09 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:55 2006