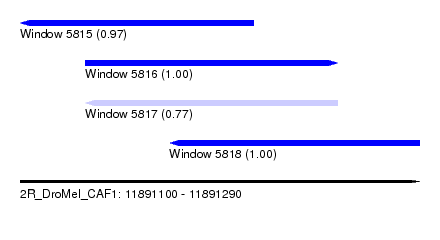
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,891,100 – 11,891,290 |
| Length | 190 |
| Max. P | 0.997619 |

| Location | 11,891,100 – 11,891,211 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.51 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -18.68 |
| Energy contribution | -20.12 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11891100 111 - 20766785 GCAGCCCCAAAUAAGGCUACAGUCAAAAUAGUGAAUAAACUACUUUAUUCAAGCUGCAAAAAUAAAUGUGAAAAAUAUUUAAUCAGAUCAGCA-AGCUGAUUACAGUC--------AAGC (((((.........(((....))).......((((((((....)))))))).))))).....(((((((.....)))))))....((((((..-..))))))......--------.... ( -25.10) >DroSec_CAF1 31610 116 - 1 GCAGCCCCAAAUAUG-CUACAGUCAAAAUAGUGAU--AACUACAUAAUUCAAGCUGCACAAGUAAAUGUGCAA-AUAUUUAAUCAGAUCAGCGGAGCUGAUUACAGUACUAUAUAAAAGC (((((......((((-.....((((......))))--.....))))......))))).....(((((((....-)))))))....(((((((...))))))).................. ( -21.50) >DroSim_CAF1 31761 119 - 1 GCAGCCCCAAAUAUGGCUACAGUCAAAAUAGUGAAUAAACUACAUUAUUCAAGCUGCACAAGUAAAUGUGCAAAAUAUUUAAUCAGAUCAGCA-AGCUGAUUACAGUCCUAUAUAAAAGC (((((........((((....))))......(((((((......))))))).))))).....(((((((.....)))))))....((((((..-..)))))).................. ( -25.20) >DroEre_CAF1 33552 119 - 1 GCAGCCCCAAACAUGGCCACAGUCAAAAUGGUGAAUAAAAUACUUUAUUCAUGCUGCAGCAGUAAAUGUACAAAAUAUUUAAUCAGAUCAGCA-AGCCGAUUACAAUCCUGUUUUACAGC (((((........((((....)))).....(((((((((....))))))))))))))((((((((((((.....)))))))....((((.(..-..).))))......)))))....... ( -24.70) >DroYak_CAF1 32050 119 - 1 GCAGCCCCAAAUAUGGCCACAGUCAAAAUAGUGAAUAAAAUACUUUAUUCAUGUUGCAGAAGUAAAUGUACAAAAAAUUUAAUCAGAUCAGCA-AGCCGAUUACAAUCCGGUUGCAAAGC ((((((........(((.............(((((((((....)))))))))((((......(((((.........))))).......)))).-.)))(((....))).))))))..... ( -23.82) >consensus GCAGCCCCAAAUAUGGCUACAGUCAAAAUAGUGAAUAAACUACUUUAUUCAAGCUGCACAAGUAAAUGUGCAAAAUAUUUAAUCAGAUCAGCA_AGCUGAUUACAGUCCUAUAUAAAAGC (((((........((((....))))......(((((((......))))))).))))).....(((((((.....)))))))....(((((((...))))))).................. (-18.68 = -20.12 + 1.44)

| Location | 11,891,131 – 11,891,251 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -26.92 |
| Energy contribution | -27.48 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11891131 120 + 20766785 AAAUAUUUUUCACAUUUAUUUUUGCAGCUUGAAUAAAGUAGUUUAUUCACUAUUUUGACUGUAGCCUUAUUUGGGGCUGCAGCAAGCGAAUUAGAUGUUGAAGAAAAUUGUCUUCCUGUA .(((.(((((((((((((..(((((....((((((((....)))))))).....(((.((((((((((....)))))))))))))))))).)))))).))))))).)))........... ( -36.90) >DroSec_CAF1 31650 116 + 1 AAAUAU-UUGCACAUUUACUUGUGCAGCUUGAAUUAUGUAGUU--AUCACUAUUUUGACUGUAG-CAUAUUUGGGGCUGCAGCAAGCGAAUUAGAUGUUGAAGAAAAUUGUCUUCCUGUA .(((((-(((...(((..((((((((((((.((.(((((((((--(.........)))))...)-)))).)).)))))))).))))..)))))))))))(((((......)))))..... ( -33.40) >DroSim_CAF1 31800 120 + 1 AAAUAUUUUGCACAUUUACUUGUGCAGCUUGAAUAAUGUAGUUUAUUCACUAUUUUGACUGUAGCCAUAUUUGGGGCUGCAGCAAGCGAAUUAGAUGUUGAAGAAAAUUGUCUUCCUGUA .(((((((((((((......))))))(((((..(((.(((((......))))).))).((((((((........))))))))))))).....)))))))(((((......)))))..... ( -36.40) >DroEre_CAF1 33591 120 + 1 AAAUAUUUUGUACAUUUACUGCUGCAGCAUGAAUAAAGUAUUUUAUUCACCAUUUUGACUGUGGCCAUGUUUGGGGCUGCAGCAAGCGAAUUAGAUGUUGAAGAAAAUUGUCUUCCUGUA .(((((((.....(((..((((((((((.((((((((....))))))))((((.......))))(((....))).)))))))).))..))).)))))))(((((......)))))..... ( -33.30) >DroYak_CAF1 32089 120 + 1 AAAUUUUUUGUACAUUUACUUCUGCAACAUGAAUAAAGUAUUUUAUUCACUAUUUUGACUGUGGCCAUAUUUGGGGCUGCAGCAAGCGAAUUAGAUGUUGAAGAAAAUUGUCUUCCUGUA .((((((((..(((((((.(((.((....((((((((....)))))))).....(((.(((..(((........)))..))))))))))).)))))))...))))))))........... ( -33.00) >consensus AAAUAUUUUGCACAUUUACUUCUGCAGCUUGAAUAAAGUAGUUUAUUCACUAUUUUGACUGUAGCCAUAUUUGGGGCUGCAGCAAGCGAAUUAGAUGUUGAAGAAAAUUGUCUUCCUGUA ...........(((((((....(((....(((((((......))))))).....(((.((((((((........))))))))))))))...))))))).(((((......)))))..... (-26.92 = -27.48 + 0.56)

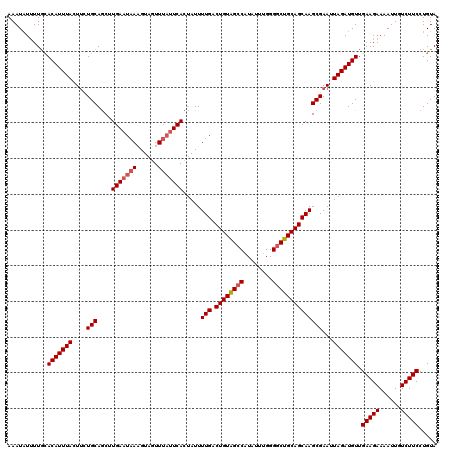
| Location | 11,891,131 – 11,891,251 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -18.68 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11891131 120 - 20766785 UACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAAGGCUACAGUCAAAAUAGUGAAUAAACUACUUUAUUCAAGCUGCAAAAAUAAAUGUGAAAAAUAUUU .....(((((......))))).........(((((((((((.((((........)))).))).))).....((((((((....))))))))................)))))........ ( -23.70) >DroSec_CAF1 31650 116 - 1 UACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAUG-CUACAGUCAAAAUAGUGAU--AACUACAUAAUUCAAGCUGCACAAGUAAAUGUGCAA-AUAUUU ((((.(((((......)))))............(((((.((((((......((((-.....((((......))))--.....))))......)))))))))))...))))...-...... ( -23.90) >DroSim_CAF1 31800 120 - 1 UACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAUGGCUACAGUCAAAAUAGUGAAUAAACUACAUUAUUCAAGCUGCACAAGUAAAUGUGCAAAAUAUUU ((((.(((((......)))))............(((((.((((((........((((....))))......(((((((......))))))).)))))))))))...)))).......... ( -28.50) >DroEre_CAF1 33591 120 - 1 UACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAACAUGGCCACAGUCAAAAUGGUGAAUAAAAUACUUUAUUCAUGCUGCAGCAGUAAAUGUACAAAAUAUUU ((((.(((((......)))))............(((.((((((((........((((....)))).....(((((((((....))))))))))))))))))))...)))).......... ( -32.70) >DroYak_CAF1 32089 120 - 1 UACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAUGGCCACAGUCAAAAUAGUGAAUAAAAUACUUUAUUCAUGUUGCAGAAGUAAAUGUACAAAAAAUUU ((((.(((((......)))))............((((.(((((((........((((....)))).....(((((((((....))))))))))))))))))))...)))).......... ( -28.00) >consensus UACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAUGGCUACAGUCAAAAUAGUGAAUAAACUACUUUAUUCAAGCUGCACAAGUAAAUGUGCAAAAUAUUU ((((.(((((......)))))..............((((((((((........((((....))))......(((((((......))))))).))))))...)))).)))).......... (-18.68 = -19.48 + 0.80)

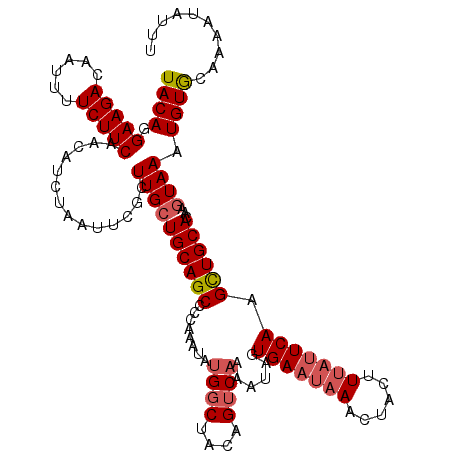
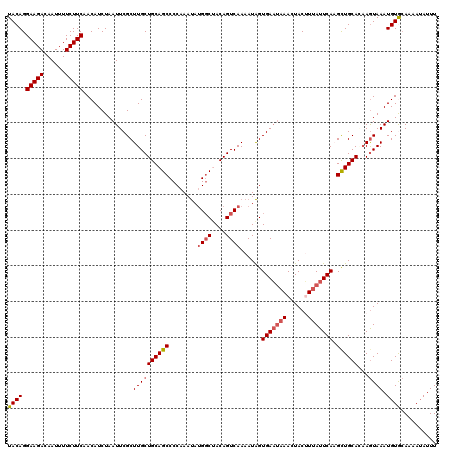
| Location | 11,891,171 – 11,891,290 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.84 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11891171 119 - 20766785 GCCUUUCCUAACUAAAACUUGAGGAAAUUAUGAAAU-UCAUACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAAGGCUACAGUCAAAAUAGUGAAUAAAC ...((((((............)))))).((((....-.))))...(((((......)))))........((((((((((((.((((........)))).))).))....))))))).... ( -24.50) >DroSec_CAF1 31689 117 - 1 GCCUUUCCUAACUAAAACUUGGGGAAAUUAUGAAAUAUCAUACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAUG-CUACAGUCAAAAUAGUGAU--AAC ...((((((((.......)))))))).........((((((....(((((......)))))..............((((((.(((.........)-)).))).)))....)))))--).. ( -20.80) >DroSim_CAF1 31840 119 - 1 GCCUUUCCUAACUAAAACUUGGGGAAAUUAUGAAAU-UCAUACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAUGGCUACAGUCAAAAUAGUGAAUAAAC ...((((((((.......))))))))..((((....-.))))...(((((......)))))........((((((((((((.((((........)))).))).))....))))))).... ( -26.30) >DroEre_CAF1 33631 119 - 1 GCCUUUCCUAACUAAAACUUGGGGAAAUUAUGAAAU-UCAUACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAACAUGGCCACAGUCAAAAUGGUGAAUAAAA ...((((((((.......))))))))..((((....-.))))...(((((......)))))........((((((((((((..(((........)))..))).))....))))))).... ( -24.30) >DroYak_CAF1 32129 119 - 1 GCCUUUCCUAACUAAAACUUGGGGAAAUUAUGAAAU-UCAUACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAUGGCCACAGUCAAAAUAGUGAAUAAAA ...((((((((.......))))))))..((((....-.))))...(((((......)))))........((((((((((((..(((........)))..))).))....))))))).... ( -24.30) >consensus GCCUUUCCUAACUAAAACUUGGGGAAAUUAUGAAAU_UCAUACAGGAAGACAAUUUUCUUCAACAUCUAAUUCGCUUGCUGCAGCCCCAAAUAUGGCUACAGUCAAAAUAGUGAAUAAAC ...((((((((.......))))))))..(((((....)))))...(((((......)))))........((((((((((((.((((........)))).))).))....))))))).... (-21.80 = -22.84 + 1.04)


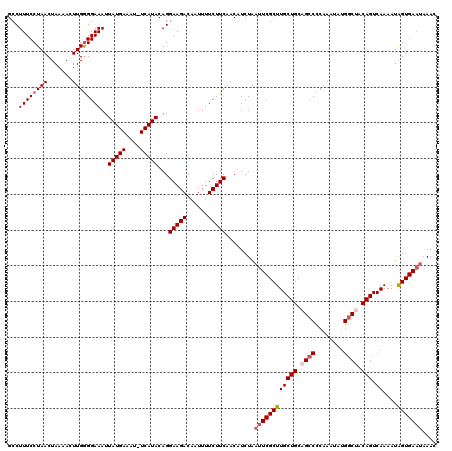
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:51 2006