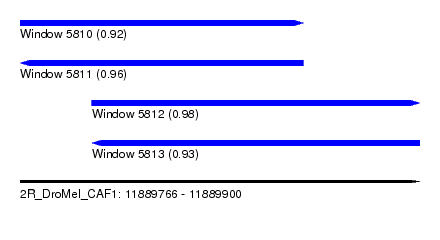
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,889,766 – 11,889,900 |
| Length | 134 |
| Max. P | 0.984877 |

| Location | 11,889,766 – 11,889,861 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -32.53 |
| Energy contribution | -32.78 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11889766 95 + 20766785 CCGUUGUGUCCACUGGCAUUGGCC--------AUUUGGGCGGUGCUGCUAUUGCUUUUGGCCCCUUUGGUUGGCCAAAUGGCAACUGGCAACGGCUGCUGCGU .(((.(((.((...(((((((.((--------....)).)))))))((((((((((((((((((...))..))))))).))))).))))...)).))).))). ( -38.90) >DroSec_CAF1 30259 94 + 1 CCGUUGUGUCCACUGGCAUUGGCC--------AUUUGGGCGGUGCUGCUAUUGCUUUUGGCCCU-UUUGUUGGCCAAAUGGCAACUGGCAACGGCUGCUGCGU .(((.(((.((...(((((((.((--------....)).)))))))((((((((((((((((..-......))))))).))))).))))...)).))).))). ( -39.30) >DroSim_CAF1 30416 94 + 1 CCGUUGUGUCCACUGGCAUUGGCC--------AUUUGGGCGGUGCUGCUAUUGCUUUUGGCCCU-UUUGUUGGCCAAAUGGCAACUGGCAACGGCUGCUGCGU .(((.(((.((...(((((((.((--------....)).)))))))((((((((((((((((..-......))))))).))))).))))...)).))).))). ( -39.30) >DroEre_CAF1 32209 102 + 1 CCGUUGUGUCCACUGCCAUUGGCCAUUUGGCCAUUUGGGCGGUGCUGGUAUUGCUUUUGGCCCU-UUUGUUGGCCAAAUGGCAACUGGCAACGGCUGUUGCGU (((((((...(((((((..(((((....)))))....)))))))......((((((((((((..-......))))))).)))))...)))))))......... ( -46.10) >consensus CCGUUGUGUCCACUGGCAUUGGCC________AUUUGGGCGGUGCUGCUAUUGCUUUUGGCCCU_UUUGUUGGCCAAAUGGCAACUGGCAACGGCUGCUGCGU (((((((.......(((((((.((............)).)))))))....((((((((((((.........))))))).)))))...)))))))......... (-32.53 = -32.78 + 0.25)



| Location | 11,889,766 – 11,889,861 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -28.22 |
| Energy contribution | -27.85 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11889766 95 - 20766785 ACGCAGCAGCCGUUGCCAGUUGCCAUUUGGCCAACCAAAGGGGCCAAAAGCAAUAGCAGCACCGCCCAAAU--------GGCCAAUGCCAGUGGACACAACGG .........(((((((..(((((..(((((((.........))))))).))))).))))).((((.....(--------(((....)))))))).......)) ( -30.70) >DroSec_CAF1 30259 94 - 1 ACGCAGCAGCCGUUGCCAGUUGCCAUUUGGCCAACAAA-AGGGCCAAAAGCAAUAGCAGCACCGCCCAAAU--------GGCCAAUGCCAGUGGACACAACGG .........(((((((..(((((..(((((((......-..))))))).))))).))))).((((.....(--------(((....)))))))).......)) ( -31.50) >DroSim_CAF1 30416 94 - 1 ACGCAGCAGCCGUUGCCAGUUGCCAUUUGGCCAACAAA-AGGGCCAAAAGCAAUAGCAGCACCGCCCAAAU--------GGCCAAUGCCAGUGGACACAACGG .........(((((((..(((((..(((((((......-..))))))).))))).))))).((((.....(--------(((....)))))))).......)) ( -31.50) >DroEre_CAF1 32209 102 - 1 ACGCAACAGCCGUUGCCAGUUGCCAUUUGGCCAACAAA-AGGGCCAAAAGCAAUACCAGCACCGCCCAAAUGGCCAAAUGGCCAAUGGCAGUGGACACAACGG .((......(((((((((...((((((((((((.....-.((((.....((.......))...))))...))))))))))))...)))))))))......)). ( -42.90) >consensus ACGCAGCAGCCGUUGCCAGUUGCCAUUUGGCCAACAAA_AGGGCCAAAAGCAAUAGCAGCACCGCCCAAAU________GGCCAAUGCCAGUGGACACAACGG .........((((((((((((((..(((((((.........))))))).))))).(((.....(((.............)))...)))...)))...)))))) (-28.22 = -27.85 + -0.37)


| Location | 11,889,790 – 11,889,900 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -42.21 |
| Energy contribution | -42.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11889790 110 + 20766785 --AUUUGGGCGGUGCUGCUAUUGCUUUUGGCCCCUUUGGUUGGCCAAAUGGCAACUGGCAACGGCUGCUGCGUUGGCCUUGUUAAAUUAUGCUCGUCAUGGCGGCACUUCCG --.....((.(((((((((((.((....((((.....))))((((((..((((.(((....))).))))...))))))................)).))))))))))).)). ( -45.30) >DroSec_CAF1 30283 109 + 1 --AUUUGGGCGGUGCUGCUAUUGCUUUUGGCCCU-UUUGUUGGCCAAAUGGCAACUGGCAACGGCUGCUGCGUUGGCCUUGUUAAAUUAUGCUCGUCAUGGCGGCACUUCCG --.....((.(((((((((((.((....(((...-((((..((((((..((((.(((....))).))))...))))))....))))....))).)).))))))))))).)). ( -44.60) >DroSim_CAF1 30440 109 + 1 --AUUUGGGCGGUGCUGCUAUUGCUUUUGGCCCU-UUUGUUGGCCAAAUGGCAACUGGCAACGGCUGCUGCGUUGGCCUUGUUAAAUUAUGCUCGUCAUGGCGGCACUUCCG --.....((.(((((((((((.((....(((...-((((..((((((..((((.(((....))).))))...))))))....))))....))).)).))))))))))).)). ( -44.60) >DroEre_CAF1 32239 111 + 1 CCAUUUGGGCGGUGCUGGUAUUGCUUUUGGCCCU-UUUGUUGGCCAAAUGGCAACUGGCAACGGCUGUUGCGUUGGCCUUGUUAAAUUAUGCUCGUCAUGGCGGCACUUCCG .......((.(((((((.((((((((((((((..-......))))))).))))).((((((.(((((......)))))))))))..............)).))))))).)). ( -39.60) >consensus __AUUUGGGCGGUGCUGCUAUUGCUUUUGGCCCU_UUUGUUGGCCAAAUGGCAACUGGCAACGGCUGCUGCGUUGGCCUUGUUAAAUUAUGCUCGUCAUGGCGGCACUUCCG .......((.(((((((((((.((....(((....((((..((((((..((((.(((....))).))))...))))))....))))....))).)).))))))))))).)). (-42.21 = -42.52 + 0.31)


| Location | 11,889,790 – 11,889,900 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -31.64 |
| Energy contribution | -31.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11889790 110 - 20766785 CGGAAGUGCCGCCAUGACGAGCAUAAUUUAACAAGGCCAACGCAGCAGCCGUUGCCAGUUGCCAUUUGGCCAACCAAAGGGGCCAAAAGCAAUAGCAGCACCGCCCAAAU-- .((..((((.((......................(((.(((((....).))))))).(((((..(((((((.........))))))).))))).)).))))..)).....-- ( -33.00) >DroSec_CAF1 30283 109 - 1 CGGAAGUGCCGCCAUGACGAGCAUAAUUUAACAAGGCCAACGCAGCAGCCGUUGCCAGUUGCCAUUUGGCCAACAAA-AGGGCCAAAAGCAAUAGCAGCACCGCCCAAAU-- .((..((((.((......................(((.(((((....).))))))).(((((..(((((((......-..))))))).))))).)).))))..)).....-- ( -33.80) >DroSim_CAF1 30440 109 - 1 CGGAAGUGCCGCCAUGACGAGCAUAAUUUAACAAGGCCAACGCAGCAGCCGUUGCCAGUUGCCAUUUGGCCAACAAA-AGGGCCAAAAGCAAUAGCAGCACCGCCCAAAU-- .((..((((.((......................(((.(((((....).))))))).(((((..(((((((......-..))))))).))))).)).))))..)).....-- ( -33.80) >DroEre_CAF1 32239 111 - 1 CGGAAGUGCCGCCAUGACGAGCAUAAUUUAACAAGGCCAACGCAACAGCCGUUGCCAGUUGCCAUUUGGCCAACAAA-AGGGCCAAAAGCAAUACCAGCACCGCCCAAAUGG .((..((((((......)).((............(((.((((((((....)))))..)))))).(((((((......-..))))))).)).......))))..))....... ( -31.90) >consensus CGGAAGUGCCGCCAUGACGAGCAUAAUUUAACAAGGCCAACGCAGCAGCCGUUGCCAGUUGCCAUUUGGCCAACAAA_AGGGCCAAAAGCAAUAGCAGCACCGCCCAAAU__ .((..((((.((.((.....((............(((.((((((((....)))))..)))))).(((((((.........))))))).)).)).)).))))..))....... (-31.64 = -31.70 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:46 2006