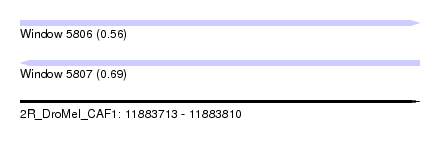
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,883,713 – 11,883,810 |
| Length | 97 |
| Max. P | 0.686518 |

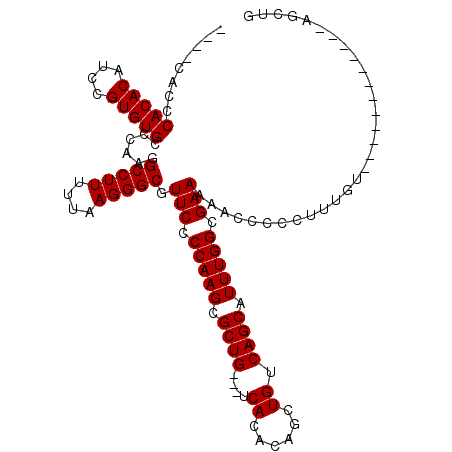
| Location | 11,883,713 – 11,883,810 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.69 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -28.66 |
| Energy contribution | -28.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11883713 97 + 20766785 CACCA------------AAAAAGGGGGUUUUUCGCCAAAUGCUGACAGCUGUGUGA---CAGCGCUUGGGGAACGCCCUUAGAAAGGCCUUGGGCACACGGUUGUGUGGGUG---- ((((.------------.....((.((....)).))........(((((((((((.---(((.((((((((....)))).....)))).)))..)))))))))))...))))---- ( -35.40) >DroSec_CAF1 24140 97 + 1 CAGCU------------ACAAAGGGGGUUUUUCGCCAAAUGCUGACAGCUGUGUGA---CAGCGCUUGGGGAACGCCCUUAAAAAGGCCUUGGGCACACGGAUGUGUGGGUG---- ((.((------------((((((((.((...((.((((.(((((.((......)).---))))).)))).)))).)))))......(((...))).........))))).))---- ( -32.70) >DroSim_CAF1 24327 97 + 1 CAGCU------------ACAAAGGGGGUUUUUCGCCAAAUGCUGACAGCUGUGUGA---CAGCGCUUGGGGAACGCCCUUAAAAAGGCCUUGGGCACACGGAUGUGUGGGUG---- ((.((------------((((((((.((...((.((((.(((((.((......)).---))))).)))).)))).)))))......(((...))).........))))).))---- ( -32.70) >DroEre_CAF1 26186 100 + 1 CAACCA-----------CCGAAGGGGG--UUUCGCCAAAUGCUGACAGCUGUGUGA---CAGCGCUUGGGGAACGCCCUUUAAAAGGCCUUGGGCACACGGAUGUGUGGGUGGGUG ...(((-----------((((((((.(--((((.((((.(((((.((......)).---))))).))))))))).)))))).............(((((....))))))))))... ( -43.60) >DroYak_CAF1 24333 114 + 1 CACCUAUGUAUGUAUGUACAAAGGGGU--UUUCGCCAAAUGCUGACAGCUGUGUGACGACAGCGCUUGGGGAACGCCCUUAAAAAGGCCUUGGGCACACGGAUGUGUGGGUGGGUG ((((((..(((.(.(((....((((((--.(((.((((.(((((....(.....)....))))).)))).))).))))))......(((...)))))).).)))..)))))).... ( -42.20) >consensus CACCU____________ACAAAGGGGGUUUUUCGCCAAAUGCUGACAGCUGUGUGA___CAGCGCUUGGGGAACGCCCUUAAAAAGGCCUUGGGCACACGGAUGUGUGGGUG____ ....................(((((.(...(((.((((.(((((....(.....)....))))).)))).)))).)))))......((((...(((((....)))))))))..... (-28.66 = -28.86 + 0.20)

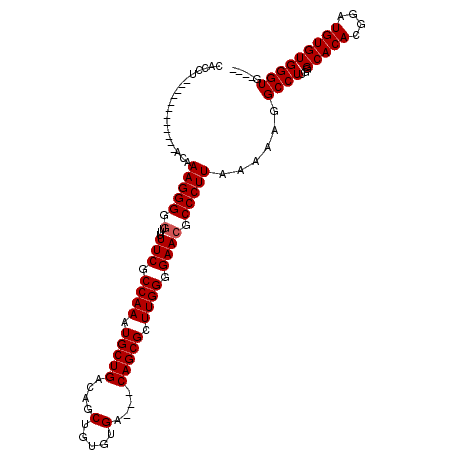
| Location | 11,883,713 – 11,883,810 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.69 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11883713 97 - 20766785 ----CACCCACACAACCGUGUGCCCAAGGCCUUUCUAAGGGCGUUCCCCAAGCGCUG---UCACACAGCUGUCAGCAUUUGGCGAAAAACCCCCUUUUU------------UGGUG ----....(((((....)))))......(((.....(((((..(((.(((((.((((---.((......)).)))).))))).))).....)))))...------------.))). ( -29.50) >DroSec_CAF1 24140 97 - 1 ----CACCCACACAUCCGUGUGCCCAAGGCCUUUUUAAGGGCGUUCCCCAAGCGCUG---UCACACAGCUGUCAGCAUUUGGCGAAAAACCCCCUUUGU------------AGCUG ----....(((((....))))).....(((..(...(((((..(((.(((((.((((---.((......)).)))).))))).))).....))))).).------------.))). ( -27.80) >DroSim_CAF1 24327 97 - 1 ----CACCCACACAUCCGUGUGCCCAAGGCCUUUUUAAGGGCGUUCCCCAAGCGCUG---UCACACAGCUGUCAGCAUUUGGCGAAAAACCCCCUUUGU------------AGCUG ----....(((((....))))).....(((..(...(((((..(((.(((((.((((---.((......)).)))).))))).))).....))))).).------------.))). ( -27.80) >DroEre_CAF1 26186 100 - 1 CACCCACCCACACAUCCGUGUGCCCAAGGCCUUUUAAAGGGCGUUCCCCAAGCGCUG---UCACACAGCUGUCAGCAUUUGGCGAAA--CCCCCUUCGG-----------UGGUUG ((.((((((((((....)))))..............(((((.(((.((((((.((((---.((......)).)))).))))).).))--).))))).))-----------))).)) ( -35.10) >DroYak_CAF1 24333 114 - 1 CACCCACCCACACAUCCGUGUGCCCAAGGCCUUUUUAAGGGCGUUCCCCAAGCGCUGUCGUCACACAGCUGUCAGCAUUUGGCGAAA--ACCCCUUUGUACAUACAUACAUAGGUG ....((((.........(((((..(((((.........(((..(((.(((((.((((.((.(.....).)).)))).))))).))).--.))))))))..))))).......)))) ( -30.59) >consensus ____CACCCACACAUCCGUGUGCCCAAGGCCUUUUUAAGGGCGUUCCCCAAGCGCUG___UCACACAGCUGUCAGCAUUUGGCGAAAAACCCCCUUUGU____________AGCUG ........(((((....)))))......(((((....))))).(((.(((((.((((....((......)).)))).))))).))).............................. (-22.14 = -22.14 + 0.00)

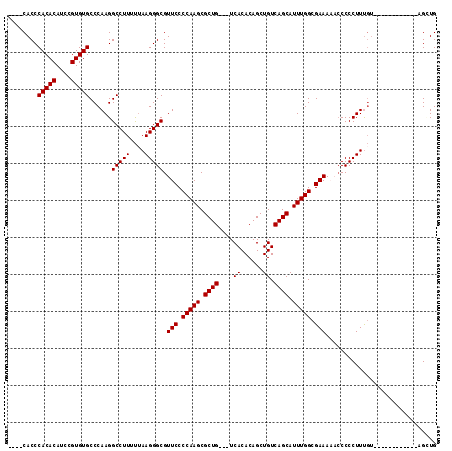
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:40 2006