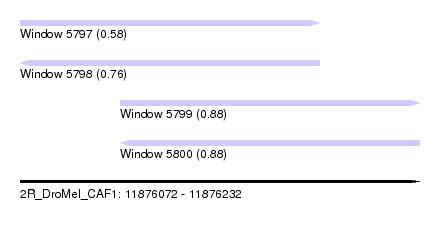
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,876,072 – 11,876,232 |
| Length | 160 |
| Max. P | 0.884100 |

| Location | 11,876,072 – 11,876,192 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -49.13 |
| Consensus MFE | -36.10 |
| Energy contribution | -36.42 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
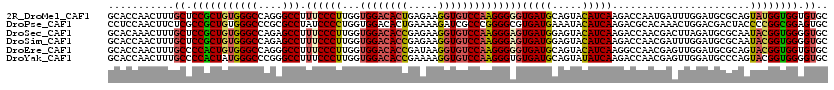
>2R_DroMel_CAF1 11876072 120 + 20766785 GCACCAACUUUGCUCCGCUGUGGGCCAGGGCCUUUCCCUUGGUGGACACUGAGAAGGUGUCCAAGGGGGUGAUGCAGUACAUCAAGACCAAUGAUUUGGAUGCGCAGUAUGGUGGUGUGC ((((((.((.((((.(((...((((....))))..(((((..((((((((.....)))))))).)))))(((((.....)))))...((((....))))..))).)))).)))))))).. ( -49.90) >DroPse_CAF1 14606 120 + 1 CCUCCAACUUCUCGCCGCUGUGGGCCCGCGCCUAUCCCCUGGUGGACACUGAAAAGAUCGCCCGGGGCGUGAUGAAAUACAUCAAGACGCACAAACUGGACGACUACCCCGGCGGAGUGC ..............((((((.(((....((((....(((.(((((...((....)).))))).)))((((((((.....))))...)))).......)).))....)))))))))..... ( -40.30) >DroSec_CAF1 16515 120 + 1 GCACAAACUUUGCUCCGCUGUGGGCCAGAGCCUUUCCCUUGGUGGACACCGAGAAGGUGUCCAAGGGAGUGAUGGAGUACAUCAAGACCAACGACUUAGAUGCGCAAUACGGUGGGGUGC ...........(((((((((((..(((..((...(((((...((((((((.....)))))))))))))))..))).(..(((((((........))).))))..)..))))))))))).. ( -48.70) >DroSim_CAF1 16102 120 + 1 GCACCAACUUUGCUCCGCUGUGGGCCAGAGCCUUUCCCUUGGUGGACACCGAGAAGGUGUCCAAGGGAGUGAUGGAGUACAUCAAGACCAACGAUUUGGAUGCGCAAUACGGUGGGGUGC ...........(((((((((((.((((...((.((((((...((((((((.....))))))))))))))(((((.....))))).............)).)).))..))))))))))).. ( -48.60) >DroEre_CAF1 18604 120 + 1 GCACCAACUUUGCCCCACUGUGGGCCAGGGCCUUUCCCUUGGUGGACACCGAUAAGGUGUCCAAGGGGGUGAUGCAGUACAUCAAGGCCAACGAGUUGGAUGCGCAGUACGGUGGUGUGC ((((((((((.((((......))))...((((((.(((((..((((((((.....)))))))).))))).((((.....))))))))))...))))))).)))(((.(((....)))))) ( -56.60) >DroYak_CAF1 16621 120 + 1 GCACCAACUUUGCCCCACUAUGGGCCCGGGCCUUUCCCUUGGUGGACACCGAAAAGGUGUCCAAGGGUGUGAUGCAGUAUAUCAAGACCAACGAGUUGGAUGCCCAGUACGGUGGGGUGC (((((.(((.((((((.....)))...((((...(((((((.((((((((.....))))))))..(((.(((((.....)))))..)))..))))..))).)))).))).)))..))))) ( -50.70) >consensus GCACCAACUUUGCUCCGCUGUGGGCCAGGGCCUUUCCCUUGGUGGACACCGAGAAGGUGUCCAAGGGAGUGAUGCAGUACAUCAAGACCAACGAGUUGGAUGCGCAGUACGGUGGGGUGC ...........((.(((((((((((....))).((((((...((((((((.....))))))))))))))(((((.....))))).......................)))))))).)).. (-36.10 = -36.42 + 0.31)
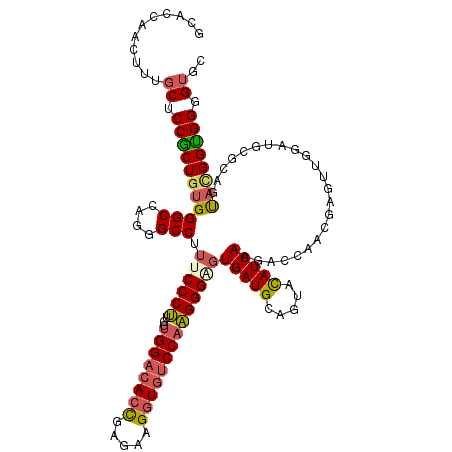
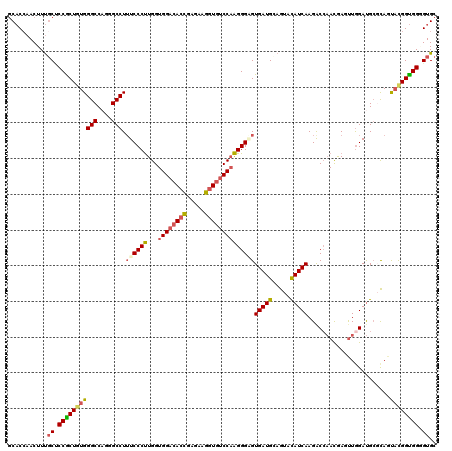
| Location | 11,876,072 – 11,876,192 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -45.75 |
| Consensus MFE | -35.89 |
| Energy contribution | -35.48 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11876072 120 - 20766785 GCACACCACCAUACUGCGCAUCCAAAUCAUUGGUCUUGAUGUACUGCAUCACCCCCUUGGACACCUUCUCAGUGUCCACCAAGGGAAAGGCCCUGGCCCACAGCGGAGCAAAGUUGGUGC ......(((((.(((..((.(((........((((((((((.....))))...(((((((((((.......)))))))...)))).))))))(((.....))).)))))..)))))))). ( -43.20) >DroPse_CAF1 14606 120 - 1 GCACUCCGCCGGGGUAGUCGUCCAGUUUGUGCGUCUUGAUGUAUUUCAUCACGCCCCGGGCGAUCUUUUCAGUGUCCACCAGGGGAUAGGCGCGGGCCCACAGCGGCGAGAAGUUGGAGG ...((((((((((((((.((..(.....)..)).))(((((.....))))).))))).)))...((((((..(((((......)))))(.(((.(.....).))).)))))))..)))). ( -45.40) >DroSec_CAF1 16515 120 - 1 GCACCCCACCGUAUUGCGCAUCUAAGUCGUUGGUCUUGAUGUACUCCAUCACUCCCUUGGACACCUUCUCGGUGUCCACCAAGGGAAAGGCUCUGGCCCACAGCGGAGCAAAGUUUGUGC ((((...((....(((((((((.(((.(....).))))))))..(((.....(((((((((((((.....))))))))...)))))..(((....)))......))))))).))..)))) ( -41.30) >DroSim_CAF1 16102 120 - 1 GCACCCCACCGUAUUGCGCAUCCAAAUCGUUGGUCUUGAUGUACUCCAUCACUCCCUUGGACACCUUCUCGGUGUCCACCAAGGGAAAGGCUCUGGCCCACAGCGGAGCAAAGUUGGUGC ((((((((.((..(((......)))..)).))).(((((((.....))))..(((((((((((((.....))))))))...))))))))((((((........))))))......))))) ( -43.20) >DroEre_CAF1 18604 120 - 1 GCACACCACCGUACUGCGCAUCCAACUCGUUGGCCUUGAUGUACUGCAUCACCCCCUUGGACACCUUAUCGGUGUCCACCAAGGGAAAGGCCCUGGCCCACAGUGGGGCAAAGUUGGUGC (((.((....))..)))(((.((((((....((((((((((.....))))...((((((((((((.....))))))))...)))).))))))...((((......))))..))))))))) ( -51.70) >DroYak_CAF1 16621 120 - 1 GCACCCCACCGUACUGGGCAUCCAACUCGUUGGUCUUGAUAUACUGCAUCACACCCUUGGACACCUUUUCGGUGUCCACCAAGGGAAAGGCCCGGGCCCAUAGUGGGGCAAAGUUGGUGC (((((........((((((..((((....))))....................((((((((((((.....))))))))...))))....))))))((((......))))......))))) ( -49.70) >consensus GCACCCCACCGUACUGCGCAUCCAAAUCGUUGGUCUUGAUGUACUGCAUCACCCCCUUGGACACCUUCUCGGUGUCCACCAAGGGAAAGGCCCUGGCCCACAGCGGAGCAAAGUUGGUGC ......(((((.(((..((.(((........((((((((((.....))))...((((((((((((.....))))))))...)))).))))))...((.....)))))))..)))))))). (-35.89 = -35.48 + -0.41)
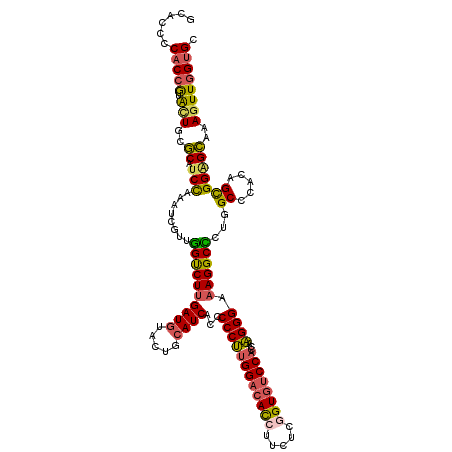

| Location | 11,876,112 – 11,876,232 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -26.95 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11876112 120 + 20766785 GGUGGACACUGAGAAGGUGUCCAAGGGGGUGAUGCAGUACAUCAAGACCAAUGAUUUGGAUGCGCAGUAUGGUGGUGUGCCGUACACGAUGAAUAAGGAGUCGGGCCAGAAUUGGGAUCA ..((((((((.....)))))))).((...(((((.....)))))...))..((((((.(((..((.(((((((.....))))))).((((.........)))).))....))).)))))) ( -39.40) >DroSec_CAF1 16555 120 + 1 GGUGGACACCGAGAAGGUGUCCAAGGGAGUGAUGGAGUACAUCAAGACCAACGACUUAGAUGCGCAAUACGGUGGGGUGCCGUACACGAUGAAUAAGGAGUCGGGCCAGAACUGGGAUCA ..((((((((.....)))))))).((...(((((.....)))))...))..((((((..((.((...((((((.....))))))..))....))...))))))..(((....)))..... ( -42.60) >DroSim_CAF1 16142 120 + 1 GGUGGACACCGAGAAGGUGUCCAAGGGAGUGAUGGAGUACAUCAAGACCAACGAUUUGGAUGCGCAAUACGGUGGGGUGCCGUACACGAUGAAUAAGGAGUCGGGCCAGAACUGGGAUCA ..((((((((.....)))))))).((...(((((.....)))))...))...((((..(.((.((..((((((.....))))))..((((.........)))).))))...)..).))). ( -39.70) >DroEre_CAF1 18644 120 + 1 GGUGGACACCGAUAAGGUGUCCAAGGGGGUGAUGCAGUACAUCAAGGCCAACGAGUUGGAUGCGCAGUACGGUGGUGUGCCGUACACGAUGAACAAGGAGUCGGGCCAGGACUGGGACCA ((((((((((.....)))))))..((...(((((.....)))))...))..(.((((...((.((.(((((((.....))))))).((((.........)))).)))).)))).).))). ( -44.20) >DroWil_CAF1 19207 117 + 1 UAUGGAGAGAAAAAAAAUAUCGAAAUCUGUAAUGGAUUAUAUCGAAUUCAAUAUGUUAGAUGACUAUCCCGGUGGAGUACCCGUAACUUUGCUCAAC---ACAAAUCAGCAAUGGGAUUA ...................(((((((((.....)))))...))))....................(((((((.(.....)))......(((((....---.......))))).))))).. ( -16.10) >DroYak_CAF1 16661 120 + 1 GGUGGACACCGAAAAGGUGUCCAAGGGUGUGAUGCAGUAUAUCAAGACCAACGAGUUGGAUGCCCAGUACGGUGGGGUGCCGUACACGAUGAACAAGGAGUCGGGCCAGGAGUGGGACCA ((((((((((.....)))))))...(((.((((.(.((.((((....((((....)))).......(((((((.....)))))))..)))).))...).)))).))).........))). ( -44.50) >consensus GGUGGACACCGAGAAGGUGUCCAAGGGAGUGAUGCAGUACAUCAAGACCAACGAGUUGGAUGCGCAGUACGGUGGGGUGCCGUACACGAUGAACAAGGAGUCGGGCCAGAACUGGGAUCA ..((((((((.....)))))))).((...(((((.....)))))...)).................(((((((.....))))))).................((.(((....)))..)). (-26.95 = -27.10 + 0.15)

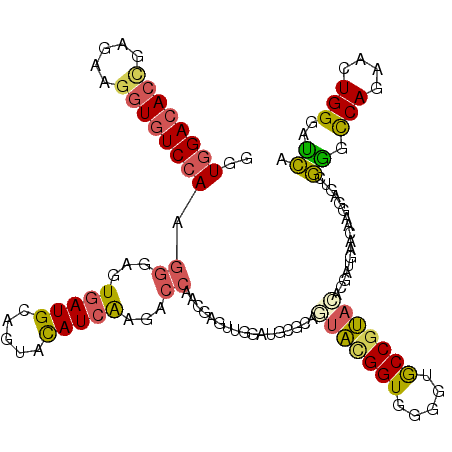

| Location | 11,876,112 – 11,876,232 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -19.87 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11876112 120 - 20766785 UGAUCCCAAUUCUGGCCCGACUCCUUAUUCAUCGUGUACGGCACACCACCAUACUGCGCAUCCAAAUCAUUGGUCUUGAUGUACUGCAUCACCCCCUUGGACACCUUCUCAGUGUCCACC .....(((((..(((...((........))..((((((.((.......)).))).)))...)))....)))))...(((((.....)))))......(((((((.......))))))).. ( -22.20) >DroSec_CAF1 16555 120 - 1 UGAUCCCAGUUCUGGCCCGACUCCUUAUUCAUCGUGUACGGCACCCCACCGUAUUGCGCAUCUAAGUCGUUGGUCUUGAUGUACUCCAUCACUCCCUUGGACACCUUCUCGGUGUCCACC .....(((....))).(((((..((((.....(((((((((.......))))).))))....))))..)))))...(((((.....)))))......((((((((.....)))))))).. ( -34.80) >DroSim_CAF1 16142 120 - 1 UGAUCCCAGUUCUGGCCCGACUCCUUAUUCAUCGUGUACGGCACCCCACCGUAUUGCGCAUCCAAAUCGUUGGUCUUGAUGUACUCCAUCACUCCCUUGGACACCUUCUCGGUGUCCACC .....(((....))).(((((...........(((((((((.......))))).))))..........)))))...(((((.....)))))......((((((((.....)))))))).. ( -30.50) >DroEre_CAF1 18644 120 - 1 UGGUCCCAGUCCUGGCCCGACUCCUUGUUCAUCGUGUACGGCACACCACCGUACUGCGCAUCCAACUCGUUGGCCUUGAUGUACUGCAUCACCCCCUUGGACACCUUAUCGGUGUCCACC .(((.........((((.(((...(((.....(((((((((.......)))))).)))....)))...)))))))..((((.....)))))))....((((((((.....)))))))).. ( -37.90) >DroWil_CAF1 19207 117 - 1 UAAUCCCAUUGCUGAUUUGU---GUUGAGCAAAGUUACGGGUACUCCACCGGGAUAGUCAUCUAACAUAUUGAAUUCGAUAUAAUCCAUUACAGAUUUCGAUAUUUUUUUUCUCUCCAUA ..(((((.(((((.(.....---..).)))))......((........)))))))...........((((((((.((..((........))..)).))))))))................ ( -21.40) >DroYak_CAF1 16661 120 - 1 UGGUCCCACUCCUGGCCCGACUCCUUGUUCAUCGUGUACGGCACCCCACCGUACUGGGCAUCCAACUCGUUGGUCUUGAUAUACUGCAUCACACCCUUGGACACCUUUUCGGUGUCCACC .(((..........(((((((.....)))......((((((.......)))))).))))..((((....))))...................)))..((((((((.....)))))))).. ( -34.50) >consensus UGAUCCCAGUUCUGGCCCGACUCCUUAUUCAUCGUGUACGGCACCCCACCGUACUGCGCAUCCAACUCGUUGGUCUUGAUGUACUCCAUCACACCCUUGGACACCUUCUCGGUGUCCACC .............((.(((((..............((((((.......))))))..............))))).))(((((.....)))))......((((((((.....)))))))).. (-19.87 = -20.66 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:33 2006