| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,874,160 – 11,874,259 |
| Length | 99 |
| Max. P | 0.998335 |

| Location | 11,874,160 – 11,874,259 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.42 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -17.85 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
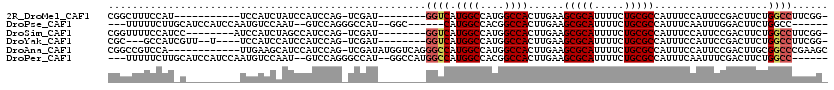
>2R_DroMel_CAF1 11874160 99 + 20766785 CGGCUUUCCAU-----------UCCAUCUAUCCAUCCAG-UCGAU--------GGUCAUGGCCAUGGCCACUUGAAGCGCAUUUUCUGCGCCAUUUCCAUUCCGACUUCUGGCCUUCGG- .((((......-----------...............((-(((((--------(((((((((....))))..))).(((((.....))))).....))))..)))))...)))).....- ( -25.85) >DroPse_CAF1 12574 101 + 1 ---UUUUUCUUGCAUCCAUCCAAUGUCCAAU--GUCCAGGGCCAU--GGC------CAUGGCCACGGCCACUUGAAGCGCAUUUUCUGCGCCAUUUCAAUUUGGACUUCUGGCC------ ---................(((..((((((.--......((((.(--(((------....)))).))))..((((((((((.....)))))...))))).))))))...)))..------ ( -35.70) >DroSim_CAF1 14192 102 + 1 CGGUUUUCCAUCC--------AUCCAUCUAGCCAUCCAG-UCGAU--------GGUCAUGGCCAUGGCCACUUGAAGCGCAUUUUCUGCGCCAUUUCCAUUCCGACUUCUGGCCUUCGG- .((....))....--------.........((((...((-(((((--------(((((((((....))))..))).(((((.....))))).....))))..)))))..))))......- ( -30.00) >DroYak_CAF1 14732 101 + 1 CGC---GCCAUCGUU--U----UCCAUCCAUCCAUCCAG-UCGAU--------GGUCAUGGCCAUGGCCACUUGAAGCGCAUUUUCUGCGCCAUUUCCAUUCCGACUUCUGGCCUUCGG- ...---((((..(..--.----..)............((-(((((--------(((((((((....))))..))).(((((.....))))).....))))..)))))..))))......- ( -25.50) >DroAna_CAF1 13220 107 + 1 CGGCCGUCCA------------UUGAAGCAUCCAUCCAG-UCGAUAUGGUCAGGGCCAUGGCCAUGGCCACUUGAAGCGCAUUUUCUGCGCCAUUUCCAUUCCGACUUGCGGCCCGAAGC .((((((...------------.((.......))...((-(((..((((((((((((((....)))))).).))).(((((.....))))).....))))..))))).))))))...... ( -38.40) >DroPer_CAF1 14286 107 + 1 ---UUUUUCUUGCAUCCAUCCAAUGUCCAAU--GUCCAGGGCCAU--GGCCAUGGCCAUGGCCACGGCCACUUGAAGCGCAUUUUCUGCGCCAUUUCAAUUUCGACUUCUGGCC------ ---................(((..(((...(--(.((..((((((--(((....)))))))))..)).)).((((((((((.....)))))...)))))....)))...)))..------ ( -37.50) >consensus CGGUUUUCCAU_C_________UCCAUCAAUCCAUCCAG_UCGAU________GGUCAUGGCCAUGGCCACUUGAAGCGCAUUUUCUGCGCCAUUUCCAUUCCGACUUCUGGCCUUCGG_ .....................................................((((.((((....))))......(((((.....)))))...................))))...... (-17.85 = -18.18 + 0.33)
| Location | 11,874,160 – 11,874,259 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.42 |
| Mean single sequence MFE | -39.03 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
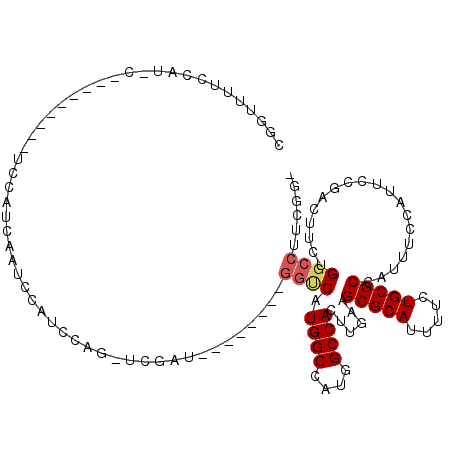
>2R_DroMel_CAF1 11874160 99 - 20766785 -CCGAAGGCCAGAAGUCGGAAUGGAAAUGGCGCAGAAAAUGCGCUUCAAGUGGCCAUGGCCAUGACC--------AUCGA-CUGGAUGGAUAGAUGGA-----------AUGGAAAGCCG -(((....(((..(((((..((((....((((((.....))))))....(((((....)))))..))--------)))))-))...))).....))).-----------..((....)). ( -33.90) >DroPse_CAF1 12574 101 - 1 ------GGCCAGAAGUCCAAAUUGAAAUGGCGCAGAAAAUGCGCUUCAAGUGGCCGUGGCCAUG------GCC--AUGGCCCUGGAC--AUUGGACAUUGGAUGGAUGCAAGAAAAA--- ------..((((..((((((........((((((.....))))))((.((.(((((((((....------)))--)))))))).)).--.)))))).))))................--- ( -41.70) >DroSim_CAF1 14192 102 - 1 -CCGAAGGCCAGAAGUCGGAAUGGAAAUGGCGCAGAAAAUGCGCUUCAAGUGGCCAUGGCCAUGACC--------AUCGA-CUGGAUGGCUAGAUGGAU--------GGAUGGAAAACCG -(((..(((((..(((((..((((....((((((.....))))))....(((((....)))))..))--------)))))-))...)))))...)))..--------((.(....).)). ( -38.70) >DroYak_CAF1 14732 101 - 1 -CCGAAGGCCAGAAGUCGGAAUGGAAAUGGCGCAGAAAAUGCGCUUCAAGUGGCCAUGGCCAUGACC--------AUCGA-CUGGAUGGAUGGAUGGA----A--AACGAUGGC---GCG -(((....(((..(((((..((((....((((((.....))))))....(((((....)))))..))--------)))))-))...))).....))).----.--.........---... ( -33.20) >DroAna_CAF1 13220 107 - 1 GCUUCGGGCCGCAAGUCGGAAUGGAAAUGGCGCAGAAAAUGCGCUUCAAGUGGCCAUGGCCAUGGCCCUGACCAUAUCGA-CUGGAUGGAUGCUUCAA------------UGGACGGCCG ......((((((.((((((.((((....((((((.....))))))(((.(.((((((....)))))))))))))).))))-)).....((....))..------------..).))))). ( -43.30) >DroPer_CAF1 14286 107 - 1 ------GGCCAGAAGUCGAAAUUGAAAUGGCGCAGAAAAUGCGCUUCAAGUGGCCGUGGCCAUGGCCAUGGCC--AUGGCCCUGGAC--AUUGGACAUUGGAUGGAUGCAAGAAAAA--- ------((((...........((((...((((((.....))))))))))(((((((((((....)))))))))--))))))(((..(--(........))..)))............--- ( -43.40) >consensus _CCGAAGGCCAGAAGUCGGAAUGGAAAUGGCGCAGAAAAUGCGCUUCAAGUGGCCAUGGCCAUGACC________AUCGA_CUGGAUGGAUAGAUCAA_________G_AUGGAAAACCG ........((((...((((....((...((((((.....))))))))..(((((....))))).............)))).))))................................... (-23.00 = -23.08 + 0.09)

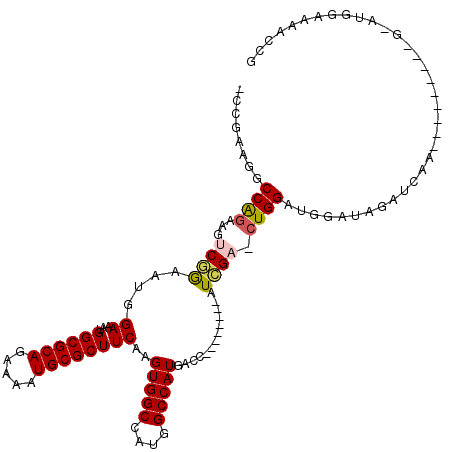
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:28 2006