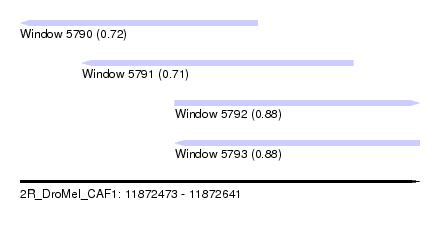
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,872,473 – 11,872,641 |
| Length | 168 |
| Max. P | 0.880516 |

| Location | 11,872,473 – 11,872,573 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.95 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11872473 100 - 20766785 AAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU-UGGAAAGUUACUACAGUUGAUAAGCAGGCGCAUAC------ACACUCGCAUUCG---CCCGCC ...((((((((..((((.(((..(((((((((......)))))).)-))..))).))))))))))))...((.((((.((.(------......).)).))---)).)). ( -27.40) >DroSec_CAF1 13046 106 - 1 AAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU-UGGAAAGUUACUACAGUUGAUAAGCGGGCGCACACACACACACACUCGCACUCA---ACCGCC ...((((((((..((((.(((..(((((((((......)))))).)-))..))).))))))))))))...(((((.(............).))))).....---...... ( -25.40) >DroSim_CAF1 12548 106 - 1 AAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU-UGGAAAGUUACUACAGUUGAUAAGCAGGCACACACACACACACACUCGCACUCA---ACCGCC ...((((((((..((((.(((..(((((((((......)))))).)-))..))).))))))))))))...((((.................)).)).....---...... ( -20.33) >DroEre_CAF1 15043 97 - 1 AAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU-UGGAAAGUUACUACAGUUGAUAAGC------------ACACACACAUACACUCGACGGCCGCC ......((((((.((((.(((..(((((((((......)))))).)-))..))).))))...(((....)))------------................)))))).... ( -20.00) >DroYak_CAF1 12943 106 - 1 AAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU-UGGAAAGUUACUACAGUUGAUAAGCUGACGCACACACACACACGCAUACACUCG---CCCGCC ...((((((((..((((.(((..(((((((((......)))))).)-))..))).))))))))))))...((....))............((........)---)..... ( -20.80) >DroAna_CAF1 11669 94 - 1 AAUUCUGCUGUCAAAAUACUUGACAUUUGAUUAAUUGAAAUCGACUUUGGAAAGUA-----AGUUGAUAAGCAGGCUGGCGCACACGGACA--------CG---CACGCC ...(((((((((((..(((((..((.((((((......))))))...))..)))))-----..))))).))))))..((((....((....--------))---..)))) ( -27.50) >consensus AAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU_UGGAAAGUUACUACAGUUGAUAAGCAGGCGCACACACACACACACUCGCACUCG___CCCGCC ...((((((((..((((.(((..((.((((((......))))))...))..))).))))))))))))........................................... (-15.36 = -15.95 + 0.59)
| Location | 11,872,499 – 11,872,613 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -12.40 |
| Energy contribution | -14.40 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11872499 114 - 20766785 CGCUUCCACUUUGGGACUGCUGCCACCCAAAUAAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU-UGGAAAGUUACUACAGUUGAUAAGCAGG .((((....((((((..........))))))............((((((((..((((.(((..(((((((((......)))))).)-))..))).)))))))))))).))))... ( -28.20) >DroSim_CAF1 12580 112 - 1 --CUUCCACUUUGGAACUGCUGCUCCCCAAAUGAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU-UGGAAAGUUACUACAGUUGAUAAGCAGG --.((((.....))))(((((((((.......)))).......((((((((..((((.(((..(((((((((......)))))).)-))..))).))))))))))))..))))). ( -31.30) >DroEre_CAF1 15069 108 - 1 CCCCUCCACU-UGGGACUGCUGCU--CCAAAUGAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU-UGGAAAGUUACUACAGUUGAUAAGC--- .(((......-.)))...((((((--(.....)))).......((((((((..((((.(((..(((((((((......)))))).)-))..))).))))))))))))..)))--- ( -26.80) >DroYak_CAF1 12975 111 - 1 CCCUUUUACU-UGGGACUGCUGCU--UCAAAUGAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU-UGGAAAGUUACUACAGUUGAUAAGCUGA (((.......-.)))...((((((--(.....)))).......((((((((..((((.(((..(((((((((......)))))).)-))..))).))))))))))))..)))... ( -26.10) >DroAna_CAF1 11693 93 - 1 --------------GAG---UUCUCCCCAAAUGAGUUAUAAAUUCUGCUGUCAAAAUACUUGACAUUUGAUUAAUUGAAAUCGACUUUGGAAAGUA-----AGUUGAUAAGCAGG --------------...---..(((.......))).........((((((((((..(((((..((.((((((......))))))...))..)))))-----..))))).))))). ( -22.00) >DroPer_CAF1 12536 93 - 1 --------------CCGUGCGACU--CCAAAUGAGUUAUAAACUCUCUUGUCAAAAUACUUGACAAUUGAUUAAUUGAAAUCGACU-UUGAAAGUA-----AGUUGAUAAGCAGG --------------((.(((((((--(.....)))))..........(((((((..(((((..(((((((((......))))))..-))).)))))-----..)))))))))))) ( -21.80) >consensus __CUUCCACU_UGGGACUGCUGCU__CCAAAUGAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAUUGAAAUCGACU_UGGAAAGUUACUACAGUUGAUAAGCAGG ..................(((..........(.(((((.....((((.((((((.....)))))).)))).))))).).(((((((.(((.......))).))))))).)))... (-12.40 = -14.40 + 2.00)

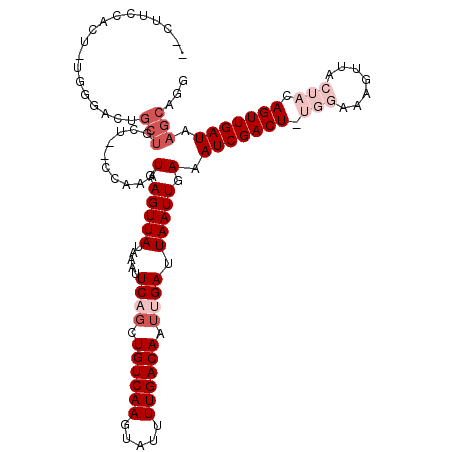
| Location | 11,872,538 – 11,872,641 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -14.87 |
| Energy contribution | -15.76 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11872538 103 + 20766785 AUUAAUCAAUUGUCAAAAUACUUGACAGCUGAAUUUAUAACUUAUUUGGGUGGCAGCAGUCCCAAAGUGGAAGCGAAGCAGGAGACAGCAGGAUGUUGCUGCG .....(((.(((((((.....))))))).))).................(..((((((.(((....((....))...((.(....).)).)))))))))..). ( -28.40) >DroSec_CAF1 13117 88 + 1 AUUAAUCAAUUGUCAAAAUACUUGACAGCUGAAUUUAUAACUCAUUUGGGGAGCAGCAGUUCCAAAGUGGAAG---------------CAGGAUGUUGCUGCG .....(((.(((((((.....))))))).))).........((((((..(((((....))))).))))))..(---------------(((.......)))). ( -24.00) >DroSim_CAF1 12619 88 + 1 AUUAAUCAAUUGUCAAAAUACUUGACAGCUGAAUUUAUAACUCAUUUGGGGAGCAGCAGUUCCAAAGUGGAAG---------------CAGGAUGUUGCUGCG .....(((.(((((((.....))))))).))).........((((((..(((((....))))).))))))..(---------------(((.......)))). ( -24.00) >DroEre_CAF1 15105 100 + 1 AUUAAUCAAUUGUCAAAAUACUUGACAGCUGAAUUUAUAACUCAUUUGG--AGCAGCAGUCCCA-AGUGGAGGGGAAACGGGCGACAGCAGGAUGUUGCUGCG .....(((.(((((((.....))))))).)))........(((((((((--..(....)..)))-))).))).(....).(((((((......)))))))... ( -29.70) >DroYak_CAF1 13014 100 + 1 AUUAAUCAAUUGUCAAAAUACUUGACAGCUGAAUUUAUAACUCAUUUGA--AGCAGCAGUCCCA-AGUAAAAGGGAAGCAGGCGACAGCAGGAUGUUGCUGCG .....(((.(((((((.....))))))).))).................--.(((((..((((.-.......))))((((.((....))....))))))))). ( -27.90) >DroPer_CAF1 12570 83 + 1 AUUAAUCAAUUGUCAAGUAUUUUGACAAGAGAGUUUAUAACUCAUUUGG--AGUCGCACGG----------------ACAGGCG--GGCAGGAUGUGUCUGCG .....((((((((((((...))))))))..((((.....))))..))))--.(((.....)----------------))..(((--((((.....))))))). ( -21.50) >consensus AUUAAUCAAUUGUCAAAAUACUUGACAGCUGAAUUUAUAACUCAUUUGG__AGCAGCAGUCCCA_AGUGGAAG_____CAGGCG__AGCAGGAUGUUGCUGCG .....(((.(((((((.....))))))).)))....................((((((((((............................))))..)))))). (-14.87 = -15.76 + 0.89)

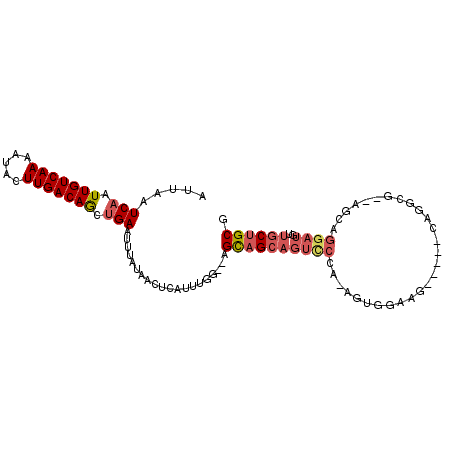

| Location | 11,872,538 – 11,872,641 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -12.90 |
| Energy contribution | -14.23 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11872538 103 - 20766785 CGCAGCAACAUCCUGCUGUCUCCUGCUUCGCUUCCACUUUGGGACUGCUGCCACCCAAAUAAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAU .((((((...(((((..((.....((...))....))..))))).))))))....................((((.(((((((...))))))).))))..... ( -21.50) >DroSec_CAF1 13117 88 - 1 CGCAGCAACAUCCUG---------------CUUCCACUUUGGAACUGCUGCUCCCCAAAUGAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAU .((((((...(((..---------------..........)))..))))))....................((((.(((((((...))))))).))))..... ( -19.70) >DroSim_CAF1 12619 88 - 1 CGCAGCAACAUCCUG---------------CUUCCACUUUGGAACUGCUGCUCCCCAAAUGAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAU .((((((...(((..---------------..........)))..))))))....................((((.(((((((...))))))).))))..... ( -19.70) >DroEre_CAF1 15105 100 - 1 CGCAGCAACAUCCUGCUGUCGCCCGUUUCCCCUCCACU-UGGGACUGCUGCU--CCAAAUGAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAU .((((((.......((....)).....((((.......-.)))).)))))).--.................((((.(((((((...))))))).))))..... ( -22.60) >DroYak_CAF1 13014 100 - 1 CGCAGCAACAUCCUGCUGUCGCCUGCUUCCCUUUUACU-UGGGACUGCUGCU--UCAAAUGAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAU .((((((......)))))).((..((.((((.......-.))))..)).)).--((((.(((((.....)))))..(((((((...))))))).))))..... ( -25.50) >DroPer_CAF1 12570 83 - 1 CGCAGACACAUCCUGCC--CGCCUGU----------------CCGUGCGACU--CCAAAUGAGUUAUAAACUCUCUUGUCAAAAUACUUGACAAUUGAUUAAU .((((.......)))).--((((...----------------..).)))(((--(.....)))).......((..(((((((.....)))))))..))..... ( -15.90) >consensus CGCAGCAACAUCCUGCU__CGCCUG_____CUUCCACU_UGGGACUGCUGCU__CCAAAUGAGUUAUAAAUUCAGCUGUCAAGUAUUUUGACAAUUGAUUAAU .((((((...(((...........................)))..))))))....................((((.((((((.....)))))).))))..... (-12.90 = -14.23 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:27 2006