
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,870,616 – 11,870,786 |
| Length | 170 |
| Max. P | 0.994975 |

| Location | 11,870,616 – 11,870,719 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.13 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -24.04 |
| Energy contribution | -24.43 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11870616 103 + 20766785 UCGUUGCCAUGUUGCCACGUUGCAUGCAAACAAGUGCAGCAAAUACACA----CACACGUACAUGCAGCGGCAACACAAAGACACGGACGCUGACACGCUGGUGCCC ..((((((..(((((...(((((((........)))))))...(((...----.....)))...)))))))))))..........((.((((........)))))). ( -30.00) >DroEre_CAF1 13181 87 + 1 UCGUUGCCAUGUUGCCACGUUGCAUGCAAACAAGUGCAGCACACACACA--------------------GGCAACACAAAGACACGGACGCUGACACGCUGGUGCCC ..((((((.(((......(((((((........)))))))......)))--------------------))))))..........((.((((........)))))). ( -25.50) >DroYak_CAF1 11031 105 + 1 UCGUUGCCAUGUUGCCACGUUGCAUGCAAACAAGUGCAGCACACAAACACACACACACUUACAGAC--UGGCAACACAAAGACACGGACGCUGACACGCUGGUGCCC ..(((....((((((((.(((((((........))))))).................(.....)..--))))))))....)))..((.((((........)))))). ( -26.60) >consensus UCGUUGCCAUGUUGCCACGUUGCAUGCAAACAAGUGCAGCACACACACA____CACAC_UACA__C___GGCAACACAAAGACACGGACGCUGACACGCUGGUGCCC ..(((....(((((((..(((((((........)))))))..................((....))...)))))))....)))..((.((((........)))))). (-24.04 = -24.43 + 0.39)

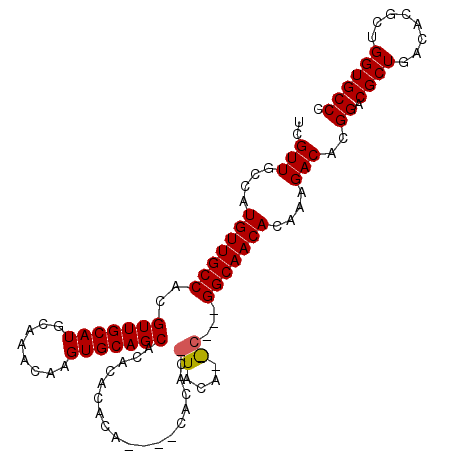
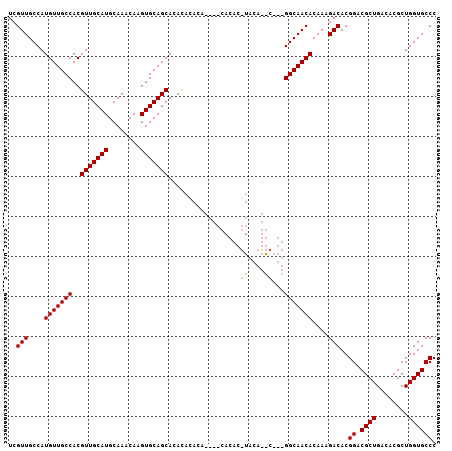
| Location | 11,870,616 – 11,870,719 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.13 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -31.13 |
| Energy contribution | -31.13 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11870616 103 - 20766785 GGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCCGCUGCAUGUACGUGUG----UGUGUAUUUGCUGCACUUGUUUGCAUGCAACGUGGCAACAUGGCAACGA ((((((..((((....))))..))))))..((((((((((((((((((((.(((----((.((....))))))).)))..)))))))..))))))))))(....).. ( -42.40) >DroEre_CAF1 13181 87 - 1 GGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCC--------------------UGUGUGUGUGCUGCACUUGUUUGCAUGCAACGUGGCAACAUGGCAACGA ((((((..((((....))))..))))))..((((((((--------------------(((.(((((((.((....))..)))))))))).))))))))(....).. ( -34.50) >DroYak_CAF1 11031 105 - 1 GGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCCA--GUCUGUAAGUGUGUGUGUGUUUGUGUGCUGCACUUGUUUGCAUGCAACGUGGCAACAUGGCAACGA ((((((..((((....))))..))))))...(((((((.--((.(((((..(.(((((..(....)..).)))).)..))))).))...(((....)))))))))). ( -36.40) >consensus GGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCC___G__UGUA_GUGUG____UGUGUGUGUGCUGCACUUGUUUGCAUGCAACGUGGCAACAUGGCAACGA ((((((..((((....))))..))))))...(((((((............................(((((((......)))).)))..(((....)))))))))). (-31.13 = -31.13 + 0.00)

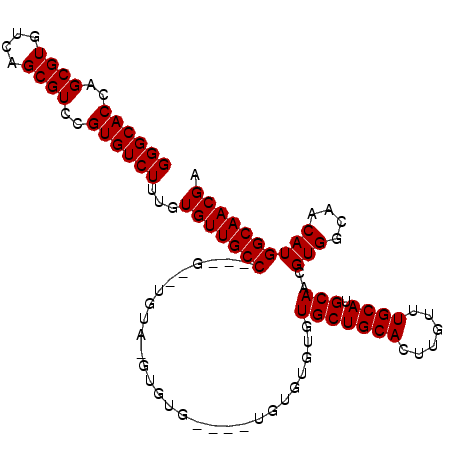
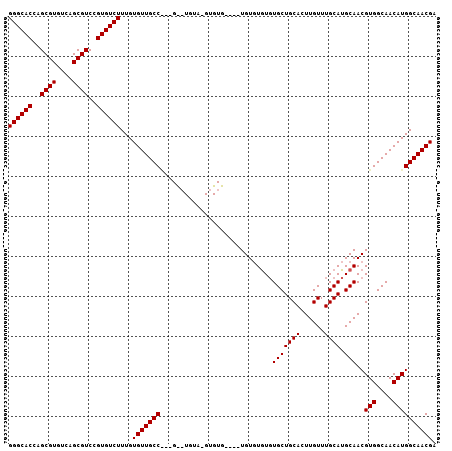
| Location | 11,870,653 – 11,870,755 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11870653 102 + 20766785 AGCAAAUACACA----CACACGUACAUGCAGCGGCAACACAAAGACACGGACGCUGACACGCUGGUGCCCGCACGUCAGCGCUCGUGCGUUUAUUUGCCGUUUCCG .(((..(((...----.....)))..)))((((((((......(.((((..(((((((..((.(....).))..)))))))..)))))......)))))))).... ( -37.80) >DroEre_CAF1 13218 86 + 1 AGCACACACACA--------------------GGCAACACAAAGACACGGACGCUGACACGCUGGUGCCCGCACGUCAGCGCUCGUGCGUUUAUUUGCCGUUUCCG ............--------------------(((((......(.((((..(((((((..((.(....).))..)))))))..)))))......)))))....... ( -30.00) >DroYak_CAF1 11068 104 + 1 AGCACACAAACACACACACACUUACAGAC--UGGCAACACAAAGACACGGACGCUGACACGCUGGUGCCCGCACGUCAGCGCUCGUGCGUUUAUUUGCCGUUUCCG .........................((((--.(((((......(.((((..(((((((..((.(....).))..)))))))..)))))......)))))))))... ( -30.70) >consensus AGCACACACACA____CACAC_UACA__C___GGCAACACAAAGACACGGACGCUGACACGCUGGUGCCCGCACGUCAGCGCUCGUGCGUUUAUUUGCCGUUUCCG ................................(((((......(.((((..(((((((..((.(....).))..)))))))..)))))......)))))....... (-29.90 = -29.90 + 0.00)


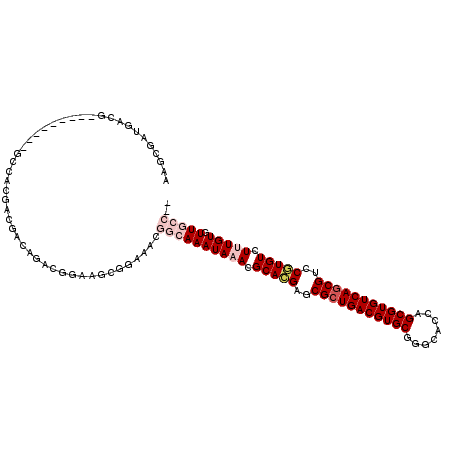
| Location | 11,870,653 – 11,870,755 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -43.43 |
| Consensus MFE | -40.13 |
| Energy contribution | -40.13 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11870653 102 - 20766785 CGGAAACGGCAAAUAAACGCACGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCCGCUGCAUGUACGUGUG----UGUGUAUUUGCU .(....)((((((((((.(((((..(((((((((((.(....).)))))))))))..))))).))))).)))))...(((.(((((....----..))))).))). ( -47.00) >DroEre_CAF1 13218 86 - 1 CGGAAACGGCAAAUAAACGCACGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCC--------------------UGUGUGUGUGCU .(....)((((((((((.(((((..(((((((((((.(....).)))))))))))..))))).))))).)))))--------------------............ ( -40.20) >DroYak_CAF1 11068 104 - 1 CGGAAACGGCAAAUAAACGCACGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCCA--GUCUGUAAGUGUGUGUGUGUUUGUGUGCU ((((...((((((((((.(((((..(((((((((((.(....).)))))))))))..))))).))))).))))).--.))))..((((..((......))..)))) ( -43.10) >consensus CGGAAACGGCAAAUAAACGCACGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCC___G__UGUA_GUGUG____UGUGUGUGUGCU .(....)((((((((((.(((((..(((((((((((.(....).)))))))))))..))))).))))).)))))................................ (-40.13 = -40.13 + -0.00)



| Location | 11,870,679 – 11,870,786 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.04 |
| Mean single sequence MFE | -43.25 |
| Consensus MFE | -30.17 |
| Energy contribution | -31.50 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11870679 107 - 20766785 CGGCGAUGACG---------GCCACGACGACAGACGGAAGCGGAAACGGCAAAUAAACGCACGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCCGC (((((((....---------(((.........(.(....))(....))))..(((((.(((((..(((((((((((.(....).)))))))))))..))))).)))))))))))). ( -48.70) >DroPse_CAF1 9032 106 - 1 AAGCGUUGAGGACGACGACGUGGACGAGGACAGACGGAA------ACGGAAAAUAAACGCAAGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUGCUUGUCUCUGUGUUG---- ..((((((.......)))))).....((.(((((((...------.))..........(((((.((((((((((((.(....).)))))))))))).))))).))))).)).---- ( -40.70) >DroEre_CAF1 13230 105 - 1 CAGCGAUGACG---------GCCACGACGACAGACGGAAGCGGAAACGGCAAAUAAACGCACGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCC-- ..(((((....---------(((.........(.(....))(....))))..(((((.(((((..(((((((((((.(....).)))))))))))..))))).)))))))))).-- ( -43.60) >DroYak_CAF1 11097 106 - 1 CAGCGAUGACG---------GCCACGACGACAGACGGAAGCGGAAACGGCAAAUAAACGCACGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCCA- ..((.....((---------....)).((.....))...))(....)((((((((((.(((((..(((((((((((.(....).)))))))))))..))))).))))).))))).- ( -43.80) >DroAna_CAF1 9249 107 - 1 AAGCGAUGACG---------CUGACGACGACAGCCGGAAACGGAAACGGCAAAUAAACGCACGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUCCUUGUCUUUGUGUUGCCGC ..(((..((((---------(.(((((.(((.((((....)(....))))........((....))((((((((((.(....).))))))))))))).)))))...)))))..))) ( -46.50) >DroPer_CAF1 10696 106 - 1 AAGCGUUGAGGACGACGACGUGGACGAGGACAGACGGAA------ACGGAAAAUAAACGCAAGAGCGGUGACGUGCGGUCACCAGCGUGUCAGCGUGCUUGUCUCUGUGUUG---- ...(((((....)))))(((.(((((((.((...((...------.))..........(((...((((((((.....)))))).)).)))....)).))))))).)))....---- ( -36.20) >consensus AAGCGAUGACG_________GCCACGACGACAGACGGAAGCGGAAACGGCAAAUAAACGCACGAGCGCUGACGUGCGGGCACCAGCGUGUCAGCGUCCGUGUCUUUGUGUUGCC__ ...............................................((((((((((.(((((..(((((((((((........)))))))))))..))))).))))).))))).. (-30.17 = -31.50 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:21 2006