| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,870,413 – 11,870,504 |
| Length | 91 |
| Max. P | 0.982555 |

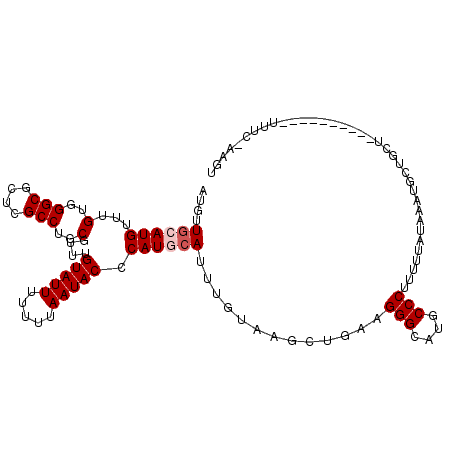
| Location | 11,870,413 – 11,870,504 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -18.47 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11870413 91 + 20766785 -----------------------GCAUUUAUAAAAAGGGCAUGCCCUACAGCUUAUAAAUGCAUGGGUAUUAAAAAAAUACAAACGCAGGCGAGCGCCCACAAACAUGCAACAU -----------------------((((((((((..((((....)))).....))))))))))(((.(((((.....)))))....((((((....)))........)))..))) ( -22.80) >DroSec_CAF1 11001 113 + 1 ACUU-GAAAUAUACUCUAGAAAAGCAUUUAUAAAAAGGGCAUGCCCUUCAGCCUAAAAAUGCAUGGGUAUUAAAAAAAUACAAACGCAGGCGAGCGCCCACAAACAUGCAACAU ....-.............................(((((....)))))...........((((((.(((((.....))))).......(((....)))......)))))).... ( -21.10) >DroSim_CAF1 10452 113 + 1 ACUU-GAAAUAUACUCUAAAAAAGCAUUUAUAAAAAGGGCAUGCCCUUCAGCCUAAAAAUGCAUGGGUAUUAAAAAAAUACAAACGCAGGCGAGCGCCCACAAACAUGCAACAU ....-.............................(((((....)))))...........((((((.(((((.....))))).......(((....)))......)))))).... ( -21.10) >DroEre_CAF1 12989 98 + 1 ---UUUAAA----------AGCAGCAU---UAAAAUGGGCAUGCCCUUAAGCUGACAAAUGAAUGGGUAUUAAAAAAAUACAAACGCAGGCGAGCGCCCACAAACAUGCAACAU ---......----------....((((---.....(((((((((((.....(........)...))))))...............((......))))))).....))))..... ( -21.60) >DroYak_CAF1 10833 101 + 1 ACUUUGUAA----------GGCAGCAU---UAAAAUGGGCAUGCCCUGAGGCUUACAAGUUAAUGGGUAUUAAAAAAAUACAAACGCAGGCGAGCGCCCACAAACAUGCAACAU .........----------....((((---.....(((((((((((...((((....))))...))))))...............((......))))))).....))))..... ( -24.50) >consensus ACUU_GAAA__________AAAAGCAUUUAUAAAAAGGGCAUGCCCUUCAGCUUAAAAAUGCAUGGGUAUUAAAAAAAUACAAACGCAGGCGAGCGCCCACAAACAUGCAACAU .......................((((.........((((((((((..................))))))...............((......))))))......))))..... (-18.47 = -18.47 + 0.00)

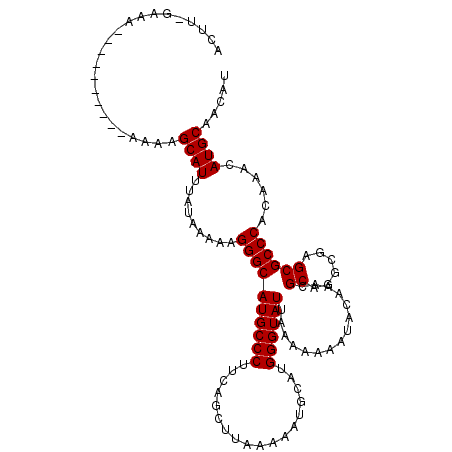

| Location | 11,870,413 – 11,870,504 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.46 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11870413 91 - 20766785 AUGUUGCAUGUUUGUGGGCGCUCGCCUGCGUUUGUAUUUUUUUAAUACCCAUGCAUUUAUAAGCUGUAGGGCAUGCCCUUUUUAUAAAUGC----------------------- .....(((((...(..(((....)))..)....(((((.....))))).)))))(((((((((....((((....)))).)))))))))..----------------------- ( -27.50) >DroSec_CAF1 11001 113 - 1 AUGUUGCAUGUUUGUGGGCGCUCGCCUGCGUUUGUAUUUUUUUAAUACCCAUGCAUUUUUAGGCUGAAGGGCAUGCCCUUUUUAUAAAUGCUUUUCUAGAGUAUAUUUC-AAGU ....((((((...(..(((....)))..)....(((((.....))))).))))))...(((....((((((....))))))...)))((((((.....)))))).....-.... ( -29.70) >DroSim_CAF1 10452 113 - 1 AUGUUGCAUGUUUGUGGGCGCUCGCCUGCGUUUGUAUUUUUUUAAUACCCAUGCAUUUUUAGGCUGAAGGGCAUGCCCUUUUUAUAAAUGCUUUUUUAGAGUAUAUUUC-AAGU ....((((((...(..(((....)))..)....(((((.....))))).))))))...(((....((((((....))))))...)))((((((.....)))))).....-.... ( -29.70) >DroEre_CAF1 12989 98 - 1 AUGUUGCAUGUUUGUGGGCGCUCGCCUGCGUUUGUAUUUUUUUAAUACCCAUUCAUUUGUCAGCUUAAGGGCAUGCCCAUUUUA---AUGCUGCU----------UUUAAA--- ..((.((((....((((((((((....((....(((((.....))))).((......))...))....))))..))))))....---)))).)).----------......--- ( -25.40) >DroYak_CAF1 10833 101 - 1 AUGUUGCAUGUUUGUGGGCGCUCGCCUGCGUUUGUAUUUUUUUAAUACCCAUUAACUUGUAAGCCUCAGGGCAUGCCCAUUUUA---AUGCUGCC----------UUACAAAGU ...(((((.(((.((((((((......))))..(((((.....))))))))).))).))))).....((((((.((........---..))))))----------))....... ( -25.50) >consensus AUGUUGCAUGUUUGUGGGCGCUCGCCUGCGUUUGUAUUUUUUUAAUACCCAUGCAUUUGUAAGCUGAAGGGCAUGCCCUUUUUAUAAAUGCUGCU__________UUUC_AAGU ....((((((...(..(((....)))..)....(((((.....))))).)))))).............(((....))).................................... (-19.86 = -20.46 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:17 2006