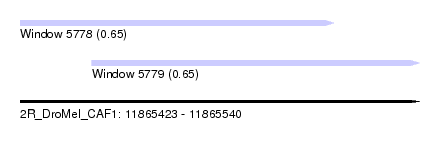
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,865,423 – 11,865,540 |
| Length | 117 |
| Max. P | 0.654404 |

| Location | 11,865,423 – 11,865,515 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -23.14 |
| Energy contribution | -22.94 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
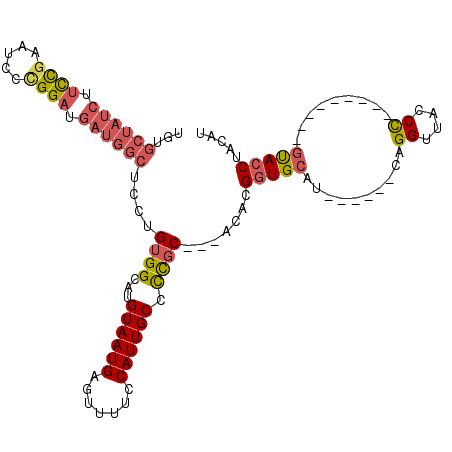
>2R_DroMel_CAF1 11865423 92 + 20766785 UGUGCUCCA-------------UCG--CU----CCACCUAUGUGCUAUCUUUCGAAUCACGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGCACACGG (((((....-------------...--..----((((....(.((((((.((((.....)))).)))))).)..))))...((((((.......))))))...)))))... ( -25.70) >DroSec_CAF1 5992 98 + 1 CGUGCUCCA-------AGUUCUCCU--CU----CCCACUAUGUGCUAUCUUUCGAAUCCCGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGCACACGG .((((....-------.........--..----.((((...(.((((((.((((.....)))).)))))).)..))))...((((((.......))))))...)))).... ( -27.60) >DroSim_CAF1 5389 105 + 1 CGUGCUCCAAGAUCCAAGCUCUCCU--CU----CCCACUAUGUGCUAUCUUUCGAAUCCCGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGCACACGG .((((..(((.....((((((....--(.----.((((...(.((((((.((((.....)))).)))))).)..))))...)....))))))....)))....)))).... ( -28.80) >DroEre_CAF1 5922 105 + 1 CCUGAUCCC------CUGCUCCCCUUGCUCCUGCCUUCUAUGUGCUAUCUUCCAAAUGCCGGAUGAUGGCUCUUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGCACACGG .........------.....((...(((...((((..(...(.((((((.(((.......))).)))))).)..).)))).((((((.......))))))...)))...)) ( -24.50) >DroYak_CAF1 5918 95 + 1 CCG---------------C-CUCUCUGCUCCUGGCUCCUAUCUGCUAUCUUCCGAAUCCCGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGCACACGG ..(---------------(-(...........)))........((((((.((((.....)))).))))))..(((((((..((((((.......))))))...)).))))) ( -29.60) >consensus CGUGCUCCA________GCUCUCCU__CU____CCCACUAUGUGCUAUCUUUCGAAUCCCGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGCACACGG ...........................................((((((.((((.....)))).))))))..(((((((..((((((.......))))))...)).))))) (-23.14 = -22.94 + -0.20)

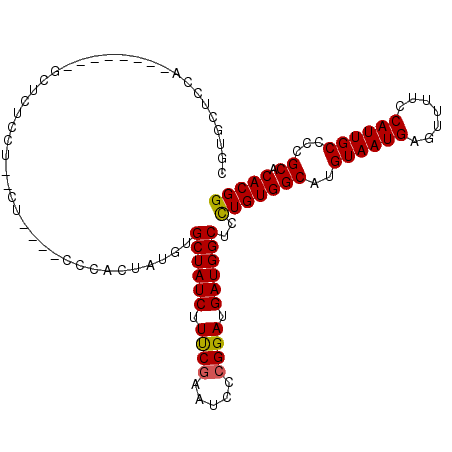
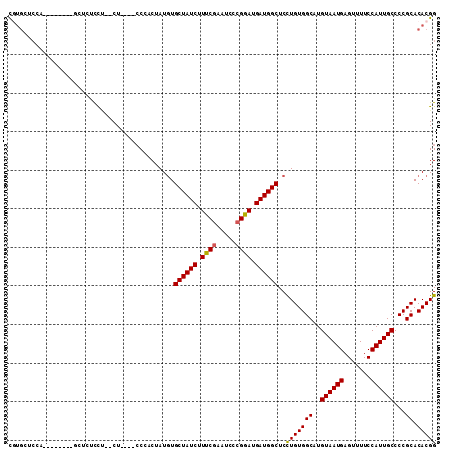
| Location | 11,865,444 – 11,865,540 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -18.71 |
| Energy contribution | -19.41 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11865444 96 + 20766785 UGUGCUAUCUUUCGAAUCACGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGC---ACACGGUGCAU------CAGGUUACCC---------GUACCUACAU (((((((((.((((.....)))).))))))..(((((((..((((((.......))))))...))---.))))).))).------.((((.....---------..)))).... ( -26.80) >DroSec_CAF1 6019 96 + 1 UGUGCUAUCUUUCGAAUCCCGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGC---ACACGGUGCAU------CAGGUAACCC---------GUACCUACAU (((((((((.((((.....)))).))))))..(((((((..((((((.......))))))...))---.))))).))).------.(((((....---------.))))).... ( -28.30) >DroSim_CAF1 5423 96 + 1 UGUGCUAUCUUUCGAAUCCCGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGC---ACACGGUGCAU------CAGGUUACCC---------GUACCUACAU (((((((((.((((.....)))).))))))..(((((((..((((((.......))))))...))---.))))).))).------.((((.....---------..)))).... ( -27.20) >DroEre_CAF1 5956 96 + 1 UGUGCUAUCUUCCAAAUGCCGGAUGAUGGCUCUUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGC---ACACGGUGCAU------CAGGUUUCCU---------GCACCUACAU .(.((((((.(((.......))).)))))).).(((((...((((((.......)))))).))))---)...((((((.------..((...)))---------)))))..... ( -30.20) >DroYak_CAF1 5942 96 + 1 UCUGCUAUCUUCCGAAUCCCGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGC---ACACGGUGCAU------CAGGUUUCCU---------GUACCUACAU ...((((((.((((.....)))).))))))...(((((...((((((.......)))))).))))---)...((((((.------..((...)))---------)))))..... ( -30.50) >DroAna_CAF1 5667 104 + 1 ----------CCUGGUUUCUGGCUCCUGGUUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCUUCCAUUACACGCUGCAUCUGCGUCAGGUUACCCCGUUUUCCAAUACCCCUGU ----------..(((....(((..(((((..(.((..((.(((((((.................))))))).))..))...)..))))).....)))....))).......... ( -23.23) >consensus UGUGCUAUCUUCCGAAUCCCGGAUGAUGGCUCCUGUGGCAUGUAAUGAGUUUUCCAUUGCCCCGC___ACACGGUGCAU______CAGGUUACCC_________GUACCUACAU ...((((((.((((.....)))).))))))....((((...((((((.......)))))).)))).......(((((..........((....)).........)))))..... (-18.71 = -19.41 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:14 2006