| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
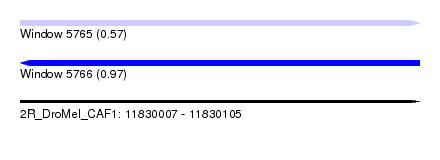
| Location | 11,830,007 – 11,830,105 |
| Length | 98 |
| Max. P | 0.966188 |

| Location | 11,830,007 – 11,830,105 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 85.58 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -18.79 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
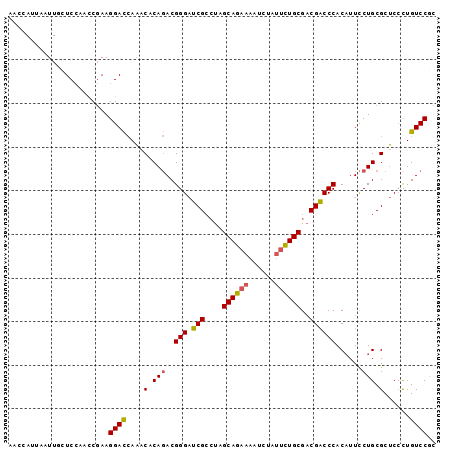
>2R_DroMel_CAF1 11830007 98 + 20766785 AACCAUUAAUUGCUCCAACCGAAGGACCAAACACAGACGGGAUCGCCUAGCAGAAAAUCUAUUCUGCGACGACCCACAUUCCUGCGCUCCCUGUCCGC .........(((.(((.......))).))).....(((((((.(((...((((((......))))))(......)........))).)))).)))... ( -23.70) >DroGri_CAF1 48818 91 + 1 AUACA----UUGUUC---UCCAAGGAUAAUGCACACACGGGCACGGGCAGCAGAAAGUCACGAUUGCGUCGUCCCACAUUUCUGCGCACGUUGUCCAC ...((----((((((---.....)))))))).......(((((((.((.(((((((((.(((((...)))))...)).))))))))).)).))))).. ( -30.50) >DroSec_CAF1 20571 98 + 1 AACCAUUAAUUGCUCCAACCGAAGGACCAAACACAGACGGGAUCGCCUAGCAGAAAAUCUAUUCUGCGACGACCCACAUUCCUGCGCUCCCUGUCCGC .........(((.(((.......))).))).....(((((((.(((...((((((......))))))(......)........))).)))).)))... ( -23.70) >DroSim_CAF1 19414 98 + 1 AACCAUUAAUUGCUCCAACCGAAGGACCAAACCCAGACGGGAUCGCCUAGCAGAAAAUCUAUUCUGCGACGACCCACAUUCCUGCGCUCCCUGUCCGC .........(((.(((.......))).))).....(((((((.(((...((((((......))))))(......)........))).)))).)))... ( -23.70) >DroEre_CAF1 20432 98 + 1 AGCCACUAAUUCCUCCAACCGAAGGACCAAACACAGACGGGAUCGCCUAGCAGAAAAUCUAUUCUGCGACGACCCACAUUCCUGCGCUCCCUGUCCGC .((.......((((.(....).)))).........(((((((.(((...((((((......))))))(......)........))).)))).))).)) ( -22.80) >DroYak_CAF1 20779 98 + 1 AGCCACUAAUUCAUCCAACCGAAGGACCAAACACAGACGGGAUCGCCUAGCAGAAAAUCUAUUCUGCGACGACCCACUUUCCUGCGCUCCCUGUCCAC .............(((.......))).........(((((((.(((...((((((......))))))(......)........))).)))).)))... ( -21.50) >consensus AACCAUUAAUUGCUCCAACCGAAGGACCAAACACAGACGGGAUCGCCUAGCAGAAAAUCUAUUCUGCGACGACCCACAUUCCUGCGCUCCCUGUCCGC .......................((((....(.(((..(((.(((....((((((......))))))..))))))......))).)......)))).. (-18.79 = -18.73 + -0.05)


| Location | 11,830,007 – 11,830,105 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.58 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -32.81 |
| Energy contribution | -33.48 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
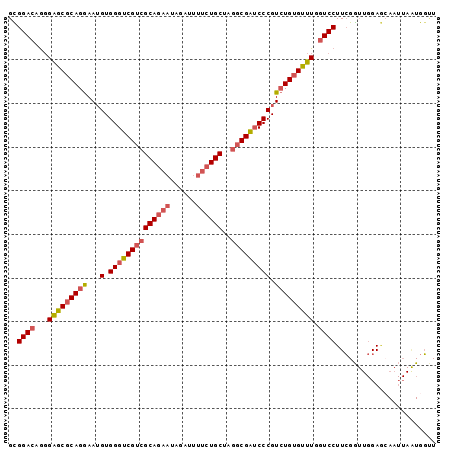
>2R_DroMel_CAF1 11830007 98 - 20766785 GCGGACAGGGAGCGCAGGAAUGUGGGUCGUCGCAGAAUAGAUUUUCUGCUAGGCGAUCCCGUCUGUGUUUGGUCCUUCGGUUGGAGCAAUUAAUGGUU ((((((...((((((((..(((.((((((((((((((......))))))..))))))))))))))))))).)))).((.....))))........... ( -38.70) >DroGri_CAF1 48818 91 - 1 GUGGACAACGUGCGCAGAAAUGUGGGACGACGCAAUCGUGACUUUCUGCUGCCCGUGCCCGUGUGUGCAUUAUCCUUGGA---GAACAA----UGUAU (.((...(((.(((((((((...(..((((.....))))..)))))))).)).))).)))....(((((((.((....))---....))----))))) ( -24.30) >DroSec_CAF1 20571 98 - 1 GCGGACAGGGAGCGCAGGAAUGUGGGUCGUCGCAGAAUAGAUUUUCUGCUAGGCGAUCCCGUCUGUGUUUGGUCCUUCGGUUGGAGCAAUUAAUGGUU ((((((...((((((((..(((.((((((((((((((......))))))..))))))))))))))))))).)))).((.....))))........... ( -38.70) >DroSim_CAF1 19414 98 - 1 GCGGACAGGGAGCGCAGGAAUGUGGGUCGUCGCAGAAUAGAUUUUCUGCUAGGCGAUCCCGUCUGGGUUUGGUCCUUCGGUUGGAGCAAUUAAUGGUU ((((((...((((.(((..(((.((((((((((((((......))))))..)))))))))))))).)))).)))).((.....))))........... ( -36.50) >DroEre_CAF1 20432 98 - 1 GCGGACAGGGAGCGCAGGAAUGUGGGUCGUCGCAGAAUAGAUUUUCUGCUAGGCGAUCCCGUCUGUGUUUGGUCCUUCGGUUGGAGGAAUUAGUGGCU ((.......((((((((..(((.((((((((((((((......))))))..)))))))))))))))))))..((((((....)))))).......)). ( -39.20) >DroYak_CAF1 20779 98 - 1 GUGGACAGGGAGCGCAGGAAAGUGGGUCGUCGCAGAAUAGAUUUUCUGCUAGGCGAUCCCGUCUGUGUUUGGUCCUUCGGUUGGAUGAAUUAGUGGCU ..((((...(((((((((...(.((((((((((((((......))))))..))))))))).))))))))).))))((((......))))......... ( -38.30) >consensus GCGGACAGGGAGCGCAGGAAUGUGGGUCGUCGCAGAAUAGAUUUUCUGCUAGGCGAUCCCGUCUGUGUUUGGUCCUUCGGUUGGAGCAAUUAAUGGUU ..((((...(((((((((...(.((((((((((((((......))))))..))))))))).))))))))).))))....................... (-32.81 = -33.48 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:01 2006